Identification of Modulators of the C. elegans Aryl Hydrocarbon Receptor and Characterization of Transcriptomic and Metabolic AhR-1 Profiles
- PMID: 35624894
- PMCID: PMC9137885
- DOI: 10.3390/antiox11051030
Identification of Modulators of the C. elegans Aryl Hydrocarbon Receptor and Characterization of Transcriptomic and Metabolic AhR-1 Profiles
Abstract
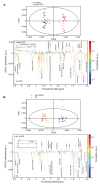
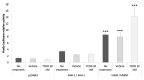
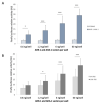
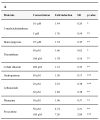
The Aryl hydrocarbon Receptor (AhR) is a xenobiotic sensor in vertebrates, regulating the metabolism of its own ligands. However, no ligand has been identified to date for any AhR in invertebrates. In C. elegans, the AhR ortholog, AHR-1, displays physiological functions. Therefore, we compared the transcriptomic and metabolic profiles of worms expressing AHR-1 or not and investigated the putative panel of chemical AHR-1 modulators. The metabolomic profiling indicated a role for AHR-1 in amino acids, carbohydrates, and fatty acids metabolism. The transcriptional profiling in neurons expressing AHR-1, identified 95 down-regulated genes and 76 up-regulated genes associated with neuronal and metabolic functions in the nervous system. A gene reporter system allowed us to identify several AHR-1 modulators including bacterial, dietary, or environmental compounds. These results shed new light on the biological functions of AHR-1 in C. elegans and perspectives on the evolution of the AhR functions across species.
Keywords: Aryl hydrocarbon Receptor; Caenorhabditis elegans; metabolomics; modulators; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

