Transcriptomal Insights of Heart Failure from Normality to Recovery
- PMID: 35625658
- PMCID: PMC9138767
- DOI: 10.3390/biom12050731
Transcriptomal Insights of Heart Failure from Normality to Recovery
Abstract
Current management of heart failure (HF) is centred on modulating the progression of symptoms and severity of left ventricular dysfunction. However, specific understandings of genetic and molecular targets are needed for more precise treatments. To attain a clearer picture of this, we studied transcriptome changes in a chronic progressive HF model. Fifteen sheep (Ovis aries) underwent supracoronary aortic banding using an inflatable cuff. Controlled and progressive induction of pressure overload in the LV was monitored by echocardiography. Endomyocardial biopsies were collected throughout the development of LV failure (LVF) and during the stage of recovery. RNA-seq data were analysed using the PANTHER database, Metascape, and DisGeNET to annotate the gene expression for functional ontologies. Echocardiography revealed distinct clinical differences between the progressive stages of hypertrophy, dilatation, and failure. A unique set of transcript expressions in each stage was identified, despite an overlap of gene expression. The removal of pressure overload allowed the LV to recover functionally. Compared to the control stage, there were a total of 256 genes significantly changed in their expression in failure, 210 genes in hypertrophy, and 73 genes in dilatation. Gene expression in the recovery stage was comparable with the control stage with a well-noted improvement in LV function. RNA-seq revealed the expression of genes in each stage that are not reported in cardiovascular pathology. We identified genes that may be potentially involved in the aetiology of progressive stages of HF, and that may provide future targets for its management.
Keywords: RNA-Seq; cardiac recovery; dilatation; heart failure; hypertrophy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
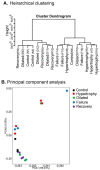


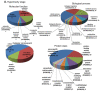




References
-
- Gerber Y., Weston S.A., Redfield M.M., Chamberlain A.M., Manemann S.M., Jiang R., Killian J.M., Roger V.L. A Contemporary Appraisal of the Heart Failure Epidemic in Olmsted County, Minnesota, 2000 to 2010. JAMA Intern. Med. 2015;175:996–1004. doi: 10.1001/jamainternmed.2015.0924. - DOI - PMC - PubMed
-
- Virani S.S., Alonso A., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Delling F.N., et al. Heart Disease and Stroke Statistics—2020 Update: A Report from the American Heart Association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. - DOI - PubMed
-
- Quttainah M., Al-Hejailan R., Saleh S., Parhar R., Conca W., Bulwer B., Moorjani N., Catarino P., Elsayed R., Shoukri M., et al. Progression of matrixin and cardiokine expression patterns in an ovine model of heart failure and recovery. Int. J. Cardiol. 2015;186:77–89. doi: 10.1016/j.ijcard.2015.03.156. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

