Utility of Bulk T-Cell Receptor Repertoire Sequencing Analysis in Understanding Immune Responses to COVID-19
- PMID: 35626377
- PMCID: PMC9140453
- DOI: 10.3390/diagnostics12051222
Utility of Bulk T-Cell Receptor Repertoire Sequencing Analysis in Understanding Immune Responses to COVID-19
Abstract
Measuring immunity to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 19 (COVID-19), can rely on antibodies, reactive T cells and other factors, with T-cell-mediated responses appearing to have greater sensitivity and longevity. Because each T cell carries an essentially unique nucleic acid sequence for its T-cell receptor (TCR), we can interrogate sequence data derived from DNA or RNA to assess aspects of the immune response. This review deals with the utility of bulk, rather than single-cell, sequencing of TCR repertoires, considering the importance of study design, in terms of cohort selection, laboratory methods and analysis. The advances in understanding SARS-CoV-2 immunity that have resulted from bulk TCR repertoire sequencing are also be discussed. The complexity of sequencing data obtained by bulk repertoire sequencing makes analysis challenging, but simple descriptive analyses, clonal analysis, searches for specific sequences associated with immune responses to SARS-CoV-2, motif-based analyses, and machine learning approaches have all been applied. TCR repertoire sequencing has demonstrated early expansion followed by contraction of SARS-CoV-2-specific clonotypes, during active infection. Maintenance of TCR repertoire diversity, including the maintenance of diversity of anti-SARS-CoV-2 response, predicts a favourable outcome. TCR repertoire narrowing in severe COVID-19 is most likely a consequence of COVID-19-associated lymphopenia. It has been possible to follow clonotypic sequences longitudinally, which has been particularly valuable for clonotypes known to be associated with SARS-CoV-2 peptide/MHC tetramer binding or with SARS-CoV-2 peptide-induced cytokine responses. Closely related clonotypes to these previously identified sequences have been shown to respond with similar kinetics during infection. A possible superantigen-like effect of the SARS-CoV-2 spike protein has been identified, by means of observing V-segment skewing in patients with severe COVID-19, together with structural modelling. Such a superantigen-like activity, which is apparently absent from other coronaviruses, may be the basis of multisystem inflammatory syndrome and cytokine storms in COVID-19. Bulk TCR repertoire sequencing has proven to be a useful and cost-effective approach to understanding interactions between SARS-CoV-2 and the human host, with the potential to inform the design of therapeutics and vaccines, as well as to provide invaluable pathogenetic and epidemiological insights.
Keywords: COVID-19; SARS-CoV-2; T-cell receptor repertoire; antibody; coronavirus; diversity; immune response; immunological memory; immunoreceptor; machine learning.
Conflict of interest statement
E.S. and A. Fowler (inventors) of GB Patent Application No: 1718238.7, for Oxford University Innovation, dated 3 November 2017; International Patent Application No: PCT/GB2018/053198 for Cambridge Enterprise, based on GB Application No: 1718238.7, dated 5 November 2018. Status: pending. The remaining authors have no conflict of interest to declare.
Figures
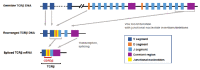
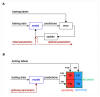

Similar articles
-
Profiling of T Cell Repertoire in SARS-CoV-2-Infected COVID-19 Patients Between Mild Disease and Pneumonia.J Clin Immunol. 2021 Aug;41(6):1131-1145. doi: 10.1007/s10875-021-01045-z. Epub 2021 May 5. J Clin Immunol. 2021. PMID: 33950324 Free PMC article.
-
Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25254-25262. doi: 10.1073/pnas.2010722117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989130 Free PMC article.
-
T cell receptor β repertoires in patients with COVID-19 reveal disease severity signatures.Front Immunol. 2023 Jul 5;14:1190844. doi: 10.3389/fimmu.2023.1190844. eCollection 2023. Front Immunol. 2023. PMID: 37475855 Free PMC article.
-
The characteristics of TCR CDR3 repertoire in COVID-19 patients and SARS-CoV-2 vaccine recipients.Virulence. 2024 Dec;15(1):2421987. doi: 10.1080/21505594.2024.2421987. Epub 2024 Nov 4. Virulence. 2024. PMID: 39468707 Free PMC article. Review.
-
Architecture of the SARS-CoV-2-specific T cell repertoire.Front Immunol. 2023 Mar 20;14:1070077. doi: 10.3389/fimmu.2023.1070077. eCollection 2023. Front Immunol. 2023. PMID: 37020560 Free PMC article. Review.
Cited by
-
T-Cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer.Int J Mol Sci. 2022 Aug 2;23(15):8590. doi: 10.3390/ijms23158590. Int J Mol Sci. 2022. PMID: 35955721 Free PMC article. Review.
-
tidytcells: standardizer for TR/MH nomenclature.Front Immunol. 2023 Oct 25;14:1276106. doi: 10.3389/fimmu.2023.1276106. eCollection 2023. Front Immunol. 2023. PMID: 37954585 Free PMC article.
-
Progress in kidney transplantation: The role for systems immunology.Front Med (Lausanne). 2022 Dec 16;9:1070385. doi: 10.3389/fmed.2022.1070385. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36590970 Free PMC article. Review.
-
T cell repertoire profiling in allografts and native tissues in recipients with COVID-19 after solid organ transplantation: Insight into T cell-mediated allograft protection from viral infection.Front Immunol. 2022 Dec 14;13:1056703. doi: 10.3389/fimmu.2022.1056703. eCollection 2022. Front Immunol. 2022. PMID: 36591281 Free PMC article.
-
The T-cell repertoire of Spanish patients with COVID-19 as a strategy to link T-cell characteristics to the severity of the disease.Hum Genomics. 2024 Sep 4;18(1):94. doi: 10.1186/s40246-024-00654-0. Hum Genomics. 2024. PMID: 39227859 Free PMC article.
References
-
- Schub D., Klemis V., Schneitler S., Mihm J., Lepper P.M., Wilkens H., Bals R., Eichler H., Gartner B.C., Becker S.L., et al. High Levels of SARS-CoV-2-Specific T Cells with Restricted Functionality in Severe Courses of COVID-19. JCI Insight. 2020;5:e142167. doi: 10.1172/jci.insight.142167. - DOI - PMC - PubMed
-
- Cheng M.H., Zhang S., Porritt R.A., Noval Rivas M., Paschold L., Willscher E., Binder M., Arditi M., Bahar I. Superantigenic Character of an Insert Unique to SARS-CoV-2 Spike Supported by Skewed TCR Repertoire in Patients with Hyperinflammation. Proc. Natl. Acad. Sci. USA. 2020;117:25254–25262. doi: 10.1073/pnas.2010722117. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

