Comprehensive Development and Implementation of Good Laboratory Practice for NGS Based Targeted Panel on Solid Tumor FFPE Tissues in Diagnostics
- PMID: 35626446
- PMCID: PMC9141409
- DOI: 10.3390/diagnostics12051291
Comprehensive Development and Implementation of Good Laboratory Practice for NGS Based Targeted Panel on Solid Tumor FFPE Tissues in Diagnostics
Abstract
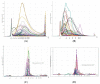
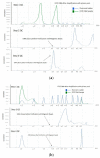
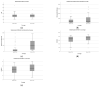
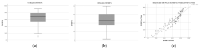
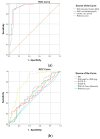
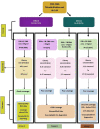
The speed, accuracy, and increasing affordability of next-generation sequencing (NGS) have revolutionized the advent of precision medicine. To date, standardized validation criteria for diagnostic accreditation do not exist due to variability across the multitude of NGS platforms and within NGS processes. In molecular diagnostics, it is necessary to ensure that the primary material of the FFPE sample has good quality and optimum quantity for the analysis, otherwise the laborious and expensive NGS test may result in unreliable information. Therefore, stringent quality control of DNA and RNA before, during, and after library preparation is an essential parameter. Considering the various challenges with the FFPE samples, we aimed to set a benchmark in QC metrics that can be utilized by molecular diagnostic laboratories for successful library preparation and high-quality NGS data output. In total, 144 DNA and 103 RNA samples of various cancer types with a maximum storage of 2 years were processed for 52 gene focus panels. During the making of DNA and RNA libraries, extensive QC check parameters were imposed at different checkpoints. The decision tree approach can be set as a benchmark for FFPE samples and as a guide to establishing a good clinical laboratory practice for targeted NGS panels.
Keywords: FFPE; diagnostics; libraries; next-generation sequencing (NGS); quality check (QC); solid tumors; standardization; targeted panel.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hovelson D.H., McDaniel A.S., Cani A.K., Johnson B., Rhodes K., Williams P.D., Bandla S., Bien G., Choppa P., Hyland F., et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17:385–399. doi: 10.1016/j.neo.2015.03.004. - DOI - PMC - PubMed
-
- Berrino E., Annaratone L., Miglio U., Maldi E., Piccinelli C., Peano E., Balmativola D., Cassoni P., Pisacane A., Sarotto I., et al. Cold Formalin Fixation Guarantees DNA Integrity in Formalin Fixed Paraffin Embedded Tissues: Premises for a Better Quality of Diagnostic and Experimental Pathology with a Specific Impact on Breast Cancer. Front. Oncol. 2020;10:173. doi: 10.3389/fonc.2020.00173. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

