Contradictory Phylogenetic Signals in the Laurasiatheria Anomaly Zone
- PMID: 35627151
- PMCID: PMC9141728
- DOI: 10.3390/genes13050766
Contradictory Phylogenetic Signals in the Laurasiatheria Anomaly Zone
Abstract
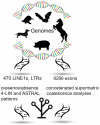
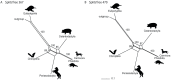
Relationships among laurasiatherian clades represent one of the most highly disputed topics in mammalian phylogeny. In this study, we attempt to disentangle laurasiatherian interordinal relationships using two independent genome-level approaches: (1) quantifying retrotransposon presence/absence patterns, and (2) comparisons of exon datasets at the levels of nucleotides and amino acids. The two approaches revealed contradictory phylogenetic signals, possibly due to a high level of ancestral incomplete lineage sorting. The positions of Eulipotyphla and Chiroptera as the first and second earliest divergences were consistent across the approaches. However, the phylogenetic relationships of Perissodactyla, Cetartiodactyla, and Ferae, were contradictory. While retrotransposon insertion analyses suggest a clade with Cetartiodactyla and Ferae, the exon dataset favoured Cetartiodactyla and Perissodactyla. Future analyses of hitherto unsampled laurasiatherian lineages and synergistic analyses of retrotransposon insertions, exon and conserved intron/intergenic sequences might unravel the conflicting patterns of relationships in this major mammalian clade.
Keywords: Laurasiatheria; Scrotifera; anomaly zone; exon coalescence; exon concatenation; retrophylogenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Feigin C.Y., Newton A.H., Doronina L., Schmitz J., Hipsley C.A., Mitchell K.J., Gower G., Llamas B., Soubrier J., Heider T.N., et al. Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nat. Ecol. Evol. 2018;2:182–192. doi: 10.1038/s41559-017-0417-y. - DOI - PubMed

