Functions and Regulation of Meiotic HORMA-Domain Proteins
- PMID: 35627161
- PMCID: PMC9141381
- DOI: 10.3390/genes13050777
Functions and Regulation of Meiotic HORMA-Domain Proteins
Abstract
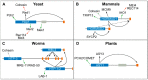
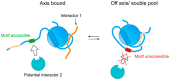
During meiosis, homologous chromosomes must recognize, pair, and recombine with one another to ensure the formation of inter-homologue crossover events, which, together with sister chromatid cohesion, promote correct chromosome orientation on the first meiotic spindle. Crossover formation requires the assembly of axial elements, proteinaceous structures that assemble along the length of each chromosome during early meiosis, as well as checkpoint mechanisms that control meiotic progression by monitoring pairing and recombination intermediates. A conserved family of proteins defined by the presence of a HORMA (HOp1, Rev7, MAd2) domain, referred to as HORMADs, associate with axial elements to control key events of meiotic prophase. The highly conserved HORMA domain comprises a flexible safety belt sequence, enabling it to adopt at least two of the following protein conformations: one closed, where the safety belt encircles a small peptide motif present within an interacting protein, causing its topological entrapment, and the other open, where the safety belt is reorganized and no interactor is trapped. Although functional studies in multiple organisms have revealed that HORMADs are crucial regulators of meiosis, the mechanisms by which HORMADs implement key meiotic events remain poorly understood. In this review, we summarize protein complexes formed by HORMADs, discuss their roles during meiosis in different organisms, draw comparisons to better characterize non-meiotic HORMADs (MAD2 and REV7), and highlight possible areas for future research.
Keywords: HORMA domain; axial element; meiosis; meiotic checkpoints; meiotic chromosomes.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

