Genome-Wide Association Analysis Reveals Genetic Architecture and Candidate Genes Associated with Grain Yield and Other Traits under Low Soil Nitrogen in Early-Maturing White Quality Protein Maize Inbred Lines
- PMID: 35627211
- PMCID: PMC9141126
- DOI: 10.3390/genes13050826
Genome-Wide Association Analysis Reveals Genetic Architecture and Candidate Genes Associated with Grain Yield and Other Traits under Low Soil Nitrogen in Early-Maturing White Quality Protein Maize Inbred Lines
Abstract
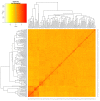
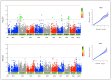
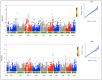
Maize production in the savannas of sub-Saharan Africa (SSA) is constrained by the low nitrogen in the soils. The identification of quantitative trait loci (QTL) conferring tolerance to low soil nitrogen (low-N) is crucial for the successful breeding of high-yielding QPM maize genotypes under low-N conditions. The objective of this study was to identify QTLs significantly associated with grain yield and other low-N tolerance-related traits under low-N. The phenotypic data of 140 early-maturing white quality protein maize (QPM) inbred lines were evaluated under low-N. The inbred lines were genotyped using 49,185 DArTseq markers, from which 7599 markers were filtered for population structure analysis and genome-wide association study (GWAS). The inbred lines were grouped into two major clusters based on the population structure analysis. The GWAS identified 24, 3, 10, and 3 significant SNPs respectively associated with grain yield, stay-green characteristic, and plant and ear aspects, under low-N. Sixteen SNP markers were physically located in proximity to 32 putative genes associated with grain yield, stay-green characteristic, and plant and ear aspects. The putative genes GRMZM2G127139, GRMZM5G848945, GRMZM2G031331, GRMZM2G003493, GRMZM2G067964, GRMZM2G180254, on chromosomes 1, 2, 8, and 10 were involved in cellular nitrogen assimilation and biosynthesis, normal plant growth and development, nitrogen assimilation, and disease resistance. Following the validation of the markers, the putative candidate genes and SNPs could be used as genomic markers for marker-assisted selection, to facilitate genetic gains for low-N tolerance in maize production.
Keywords: GWAS; QPM; candidate genes; low-N; marker-assisted selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Hubert B., Rosegrant M., Van Boekel M.A., Ortiz R. The future of food: Scenarios for 2050. Crop Sci. 2010;50:S33. doi: 10.2135/cropsci2009.09.0530. - DOI
-
- Shiferaw B., Prasanna B.M., Hellin J., Bänziger M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Sec. 2011;3:307–327. doi: 10.1007/s12571-011-0140-5. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

