Comparing and Optimizing RNA Extraction from the Pancreas of Diabetic and Healthy Rats for Gene Expression Analyses
- PMID: 35627265
- PMCID: PMC9141713
- DOI: 10.3390/genes13050881
Comparing and Optimizing RNA Extraction from the Pancreas of Diabetic and Healthy Rats for Gene Expression Analyses
Abstract
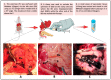
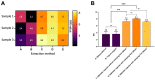
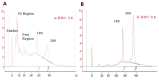
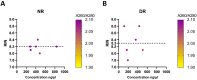
Advanced differential gene expression analysis requires high-quality RNA. However, isolating intact pancreatic RNA is challenging due to abundant pancreatic ribonucleases, which limits efficient downstream gene expression analysis. RNAlater treatment reduces endogenous ribonucleases effects through either pre-organ excision via organ mass or bile duct direct injection or organ mass injection post-isolation. We compared RNA extraction protocols to establish a reproducible and effective pancreatic RNA extraction method to obtain high RNA integrity number (RIN) values from healthy and streptozotocin (STZ)-induced diabetic rats for gene expression analyses. Different methods were tested focusing on RNase activity inhibition using RNAlater (Qiagen) pre-harvest of the pancreatic tissue, and extracted RNA quality and concentration were analyzed using NanoDrop spectrophotometer, Agilent Bioanalyzer, and RT-PCR. Inclusion of several pre- and post-excision modifications in the RNeasy Mini Kit (Qiagen) protocol resulted in RIN values more than two-fold higher compared to those using the standard protocol. Additionally, RT-PCR amplification of the housekeeping gene, β-actin, revealed no differences in extracted RNA quality from healthy and STZ-induced diabetic rats. We compared and developed a more effective and reproducible pancreatic RNA extraction method from healthy and diabetic rats, which resulted in RNA of superior quality and integrity and is suitable for complex molecular investigations.
Keywords: RIN value; RNA extraction; RNA isolation; STZ-diabetic rat model; pancreas.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


Similar articles
-
Optimization of RNA extraction from rat pancreatic tissue.Iran J Med Sci. 2014 May;39(3):282-8. Iran J Med Sci. 2014. PMID: 24850986 Free PMC article.
-
A universal method for high-quality RNA extraction from plant tissues rich in starch, proteins and fiber.Sci Rep. 2020 Oct 9;10(1):16887. doi: 10.1038/s41598-020-73958-5. Sci Rep. 2020. PMID: 33037299 Free PMC article.
-
High-quality RNA extraction from copepods for Next Generation Sequencing: A comparative study.Mar Genomics. 2015 Dec;24 Pt 1:115-8. doi: 10.1016/j.margen.2014.12.004. Epub 2014 Dec 26. Mar Genomics. 2015. PMID: 25546577
-
Extraction of high-quality RNA from mouse pancreatic tumors.MethodsX. 2020 Nov 26;7:101163. doi: 10.1016/j.mex.2020.101163. eCollection 2020. MethodsX. 2020. PMID: 33665149 Free PMC article.
-
Nigella sativa and thymoquinone suppress cyclooxygenase-2 and oxidative stress in pancreatic tissue of streptozotocin-induced diabetic rats.Pancreas. 2013 Jul;42(5):841-9. doi: 10.1097/MPA.0b013e318279ac1c. Pancreas. 2013. PMID: 23429494
Cited by
-
Optimisation of Sample Preparation from Primary Mouse Tissue to Maintain RNA Integrity for Methods Examining Translational Control.Cancers (Basel). 2023 Aug 5;15(15):3985. doi: 10.3390/cancers15153985. Cancers (Basel). 2023. PMID: 37568801 Free PMC article.
-
Identification of spatially-resolved markers of malignant transformation in Intraductal Papillary Mucinous Neoplasms.Nat Commun. 2024 Mar 29;15(1):2764. doi: 10.1038/s41467-024-46994-2. Nat Commun. 2024. PMID: 38553466 Free PMC article.
-
Interleukin 21-Armed EGFR-VHH-CAR-T Cell Therapy for the Treatment of Esophageal Squamous Cell Carcinoma.Biomedicines. 2025 Jun 30;13(7):1598. doi: 10.3390/biomedicines13071598. Biomedicines. 2025. PMID: 40722671 Free PMC article.
-
Ginger extract promotes pancreatic islets regeneration in streptozotocin-induced diabetic rats.Biosci Rep. 2025 Mar 13;45(3):BSR20241510. doi: 10.1042/BSR20241510. Biosci Rep. 2025. PMID: 40014427 Free PMC article.
-
Among Other Tissues, Short-Term Garlic Oral Treatment Incrementally Improves Indicants of Only Pancreatic Islets of Langerhans Histology and Insulin mRNA Transcription and Synthesis in Diabetic Rats.Biology (Basel). 2024 May 18;13(5):355. doi: 10.3390/biology13050355. Biology (Basel). 2024. PMID: 38785837 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

