Brain Proteome and Behavioural Analysis in Wild Type, BDNF+/- and BDNF-/- Adult Zebrafish (Danio rerio) Exposed to Two Different Temperatures
- PMID: 35628418
- PMCID: PMC9146406
- DOI: 10.3390/ijms23105606
Brain Proteome and Behavioural Analysis in Wild Type, BDNF+/- and BDNF-/- Adult Zebrafish (Danio rerio) Exposed to Two Different Temperatures
Abstract
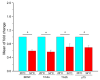
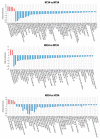
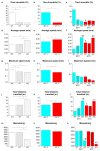
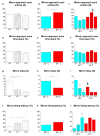
Experimental evidence suggests that environmental stress conditions can alter the expression of BDNF and that the expression of this neurotrophin influences behavioural responses in mammalian models. It has been recently demonstrated that exposure to 34 °C for 21 days alters the brain proteome and behaviour in zebrafish. The aim of this work was to investigate the role of BDNF in the nervous system of adult zebrafish under control and heat treatment conditions. For this purpose, zebrafish from three different genotypes (wild type, heterozygous BDNF+/- and knock out BDNF-/-) were kept for 21 days at 26 °C or 34 °C and then euthanized for brain molecular analyses or subjected to behavioural tests (Y-maze test, novel tank test, light and dark test, social preference test, mirror biting test) for assessing behavioural aspects such as boldness, anxiety, social preference, aggressive behaviour, interest for the novel environment and exploration. qRT-PCR analysis showed the reduction of gene expression of BDNF and its receptors after heat treatment in wild type zebrafish. Moreover, proteomic analysis and behavioural tests showed genotype- and temperature-dependent effects on brain proteome and behavioural responding. Overall, the absent expression of BDNF in KO alters (1) the brain proteome by reducing the expression of proteins involved in synapse functioning and neurotransmitter-mediated transduction; (2) the behaviour, which can be interpreted as bolder and less anxious and (3) the cellular and behavioural response to thermal treatment.
Keywords: BDNF; behaviour; proteomic; temperature; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










References
-
- Nittoli V., Sepe R.M., Coppola U., D’Agostino Y., De Felice E., Palladino A., Vassalli Q.A., Locascio A., Ristoratore F., Spagnuolo A., et al. A comprehensive analysis of neurotrophins and neurotrophin tyrosine kinase receptors expression during development of zebrafish. J. Comp. Neurol. 2018;526:1057–1072. doi: 10.1002/cne.24391. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

