Penicillin-Binding Proteins, β-Lactamases, and β-Lactamase Inhibitors in β-Lactam-Producing Actinobacteria: Self-Resistance Mechanisms
- PMID: 35628478
- PMCID: PMC9146315
- DOI: 10.3390/ijms23105662
Penicillin-Binding Proteins, β-Lactamases, and β-Lactamase Inhibitors in β-Lactam-Producing Actinobacteria: Self-Resistance Mechanisms
Abstract
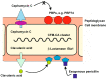
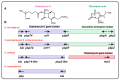
The human society faces a serious problem due to the widespread resistance to antibiotics in clinical practice. Most antibiotic biosynthesis gene clusters in actinobacteria contain genes for intrinsic self-resistance to the produced antibiotics, and it has been proposed that the antibiotic resistance genes in pathogenic bacteria originated in antibiotic-producing microorganisms. The model actinobacteria Streptomyces clavuligerus produces the β-lactam antibiotic cephamycin C, a class A β-lactamase, and the β lactamases inhibitor clavulanic acid, all of which are encoded in a gene supercluster; in addition, it synthesizes the β-lactamase inhibitory protein BLIP. The secreted clavulanic acid has a synergistic effect with the cephamycin produced by the same strain in the fight against competing microorganisms in its natural habitat. High levels of resistance to cephamycin/cephalosporin in actinobacteria are due to the presence (in their β-lactam clusters) of genes encoding PBPs which bind penicillins but not cephalosporins. We have revised the previously reported cephamycin C and clavulanic acid gene clusters and, in addition, we have searched for novel β-lactam gene clusters in protein databases. Notably, in S. clavuligerus and Nocardia lactamdurans, the β-lactamases are retained in the cell wall and do not affect the intracellular formation of isopenicillin N/penicillin N. The activity of the β-lactamase in S. clavuligerus may be modulated by the β-lactamase inhibitory protein BLIP at the cell-wall level. Analysis of the β-lactam cluster in actinobacteria suggests that these clusters have been moved by horizontal gene transfer between different actinobacteria and have culminated in S. clavuligerus with the organization of an elaborated set of genes designed for fine tuning of antibiotic resistance and cell wall remodeling for the survival of this Streptomyces species. This article is focused specifically on the enigmatic connection between β-lactam biosynthesis and β-lactam resistance mechanisms in the producer actinobacteria.
Keywords: Streptomyces clavuligerus; antibiotic resistance; cephamycin C; clavulanic acid; clusters organization and evolution; penicillin-binding proteins; superclusters; β-lactamases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genes for a beta-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans.EMBO J. 1993 Feb;12(2):631-9. doi: 10.1002/j.1460-2075.1993.tb05696.x. EMBO J. 1993. PMID: 8440253 Free PMC article.
-
Penicillin-binding proteins in Actinobacteria.J Antibiot (Tokyo). 2015 Apr;68(4):223-45. doi: 10.1038/ja.2014.148. Epub 2014 Oct 29. J Antibiot (Tokyo). 2015. PMID: 25351947 Review.
-
New aspects of genes and enzymes for beta-lactam antibiotic biosynthesis.Appl Microbiol Biotechnol. 1998 Jul;50(1):1-15. doi: 10.1007/s002530051249. Appl Microbiol Biotechnol. 1998. PMID: 9720195 Review.
-
Construction and analysis of ss-lactamase-inhibitory protein (BLIP) non-producer mutants of Streptomyces clavuligerus.Microbiology (Reading). 2001 Feb;147(Pt 2):325-335. doi: 10.1099/00221287-147-2-325. Microbiology (Reading). 2001. PMID: 11158349
-
Genes for beta-lactam antibiotic biosynthesis.Antonie Van Leeuwenhoek. 1995;67(2):181-200. doi: 10.1007/BF00871213. Antonie Van Leeuwenhoek. 1995. PMID: 7771766 Review.
Cited by
-
Ultrasonic-Assisted Synthesis of Heterocyclic Curcumin Analogs as Antidiabetic, Antibacterial, and Antioxidant Agents Combined with in vitro and in silico Studies.Adv Appl Bioinform Chem. 2023 Jul 28;16:61-91. doi: 10.2147/AABC.S403413. eCollection 2023. Adv Appl Bioinform Chem. 2023. PMID: 37533689 Free PMC article.
-
Interconnected Set of Enzymes Provide Lysine Biosynthetic Intermediates and Ornithine Derivatives as Key Precursors for the Biosynthesis of Bioactive Secondary Metabolites.Antibiotics (Basel). 2023 Jan 12;12(1):159. doi: 10.3390/antibiotics12010159. Antibiotics (Basel). 2023. PMID: 36671360 Free PMC article. Review.
-
Virulence and Antibiotic Resistance Genes in Enterococcus from Wastewater for Reuse and Their Health Impact.Microorganisms. 2025 Apr 30;13(5):1045. doi: 10.3390/microorganisms13051045. Microorganisms. 2025. PMID: 40431216 Free PMC article.
-
Advance Screening of Bis-azetidinone Derivatives: Synthesis, Spectroscopy, Antioxidant and Antimicrobial Analysis with Molecular Docking Assessment.Curr Org Synth. 2025;22(3):396-409. doi: 10.2174/0115701794318870240923073910. Curr Org Synth. 2025. PMID: 40259592
-
Facile Synthesis of 5-Bromo-N-Alkylthiophene-2-Sulfonamides and Its Activities Against Clinically Isolated New Delhi Metallo-β-Lactamase Producing Klebsiella pneumoniae ST147.Infect Drug Resist. 2024 Jul 11;17:2943-2955. doi: 10.2147/IDR.S455979. eCollection 2024. Infect Drug Resist. 2024. PMID: 39011342 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

