The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia fructicola
- PMID: 35628739
- PMCID: PMC9147972
- DOI: 10.3390/jof8050481
The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia fructicola
Abstract
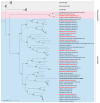
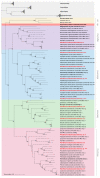
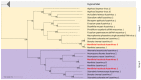
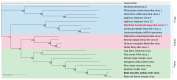
The fungus Monilinia fructicola is responsible for brown rot on stone and pome fruit and causes heavy yield losses both pre- and post-harvest. Several mycoviruses are known to infect fungal plant pathogens. In this study, a metagenomic approach was applied to obtain a comprehensive characterization of the mycovirome in a worldwide collection of 58 M. fructicola strains. Deep sequencing of double-stranded (ds)RNA extracts revealed a great abundance and variety of mycoviruses. A total of 32 phylogenetically distinct positive-sense (+) single-stranded (ss)RNA viruses were identified. They included twelve mitoviruses, one in the proposed family Splipalmiviridae, and twelve botourmiaviruses (phylum Lenarviricota), eleven of which were novel viral species; two hypoviruses, three in the proposed family Fusariviridae, and one barnavirus (phylum Pisuviricota); as well as one novel beny-like virus (phylum Kitrinoviricota), the first one identified in Ascomycetes. A partial sequence of a new putative ssDNA mycovirus related to viruses within the Parvoviridae family was detected in a M. fructicola isolate from Serbia. The availability of genomic sequences of mycoviruses will serve as a solid basis for further research aimed at deepening the knowledge on virus-host and virus-virus interactions and to explore their potential as biocontrol agents against brown rot disease.
Keywords: +ssRNA virus; barnavirus; benyvirus; botourmiavirus; fusarivirus; hypovirus; mitovirus; mycovirus; parvovirus; splipalmivirus; stone fruit.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Uchida K., Sakuta K., Ito A., Takahashi Y., Katayama Y., Omatsu T., Mizutani T., Arie T., Komatsu K., Fukuhara T., et al. Two novel endornaviruses co-infecting a Phytophthora pathogen of Asparagus officinalis modulate the developmental stages and fungicide sensitivities of the host oomycete. Front. Microbiol. 2021;12:122. doi: 10.3389/fmicb.2021.633502. - DOI - PMC - PubMed
-
- Sutela S., Forgia M., Vainio E.J., Chiapello M., Daghino S., Vallino M., Martino E., Girlanda M., Perotto S., Turina M. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol. 2020;6:veaa076. doi: 10.1093/ve/veaa076. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

