An Unprecedented Number of Cytochrome P450s Are Involved in Secondary Metabolism in Salinispora Species
- PMID: 35630316
- PMCID: PMC9143469
- DOI: 10.3390/microorganisms10050871
An Unprecedented Number of Cytochrome P450s Are Involved in Secondary Metabolism in Salinispora Species
Abstract
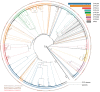
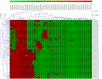
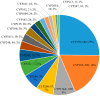
Cytochrome P450 monooxygenases (CYPs/P450s) are heme thiolate proteins present in species across the biological kingdoms. By virtue of their broad substrate promiscuity and regio- and stereo-selectivity, these enzymes enhance or attribute diversity to secondary metabolites. Actinomycetes species are well-known producers of secondary metabolites, especially Salinispora species. Despite the importance of P450s, a comprehensive comparative analysis of P450s and their role in secondary metabolism in Salinispora species is not reported. We therefore analyzed P450s in 126 strains from three different species Salinispora arenicola, S. pacifica, and S. tropica. The study revealed the presence of 2643 P450s that can be grouped into 45 families and 103 subfamilies. CYP107 and CYP125 families are conserved, and CYP105 and CYP107 families are bloomed (a P450 family with many members) across Salinispora species. Analysis of P450s that are part of secondary metabolite biosynthetic gene clusters (smBGCs) revealed Salinispora species have an unprecedented number of P450s (1236 P450s-47%) part of smBGCs compared to other bacterial species belonging to the genera Streptomyces (23%) and Mycobacterium (11%), phyla Cyanobacteria (8%) and Firmicutes (18%) and the classes Alphaproteobacteria (2%) and Gammaproteobacteria (18%). A peculiar characteristic of up to six P450s in smBGCs was observed in Salinispora species. Future characterization Salinispora species P450s and their smBGCs have the potential for discovering novel secondary metabolites.
Keywords: Mycobacterium; Salinispora arenicola; Streptomyces; actinomycete; biosynthetic gene clusters; cytochrome P450; diversity; genome-data mining; marine; natural products; secondary metabolites.
Conflict of interest statement
The authors declare no conflict of interest, and the funders had no role in the study’s design, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

