Hit-to-Lead Short Peptides against Dengue Type 2 Envelope Protein: Computational and Experimental Investigations
- PMID: 35630712
- PMCID: PMC9146555
- DOI: 10.3390/molecules27103233
Hit-to-Lead Short Peptides against Dengue Type 2 Envelope Protein: Computational and Experimental Investigations
Abstract
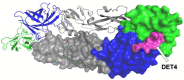
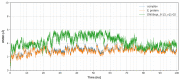
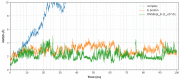
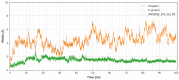
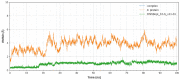
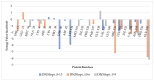
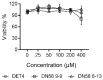
Data from the World Health Organisation show that the global incidence of dengue infection has risen drastically, with an estimated 400 million cases of dengue infection occurring annually. Despite this worrying trend, there is still no therapeutic treatment available. Herein, we investigated short peptide fragments with a varying total number of amino acid residues (peptide fragments) from previously reported dengue virus type 2 (DENV2) peptide-based inhibitors, DN58wt (GDSYIIIGVEPGQLKENWFKKGSSIGQMF), DN58opt (TWWCFYFCRRHHPFWFFYRHN), DS36wt (LITVNPIVTEKDSPVNIEAE), and DS36opt (RHWEQFYFRRRERKFWLFFW), aided by in silico approaches: peptide-protein molecular docking and 100 ns of molecular dynamics (MD) simulation via molecular mechanics using Poisson-Boltzmann surface area (MMPBSA) and molecular mechanics generalised Born surface area (MMGBSA) methods. A library of 11,699 peptide fragments was generated, subjected to in silico calculation, and the candidates with the excellent binding affinity and shown to be stable in the DI-DIII binding pocket of DENV2 envelope (E) protein were determined. Selected peptides were synthesised using conventional Fmoc solid-phase peptide chemistry, purified by RP-HPLC, and characterised using LCMS. In vitro studies followed, to test for the peptides' toxicity and efficacy in inhibiting the DENV2 growth cycle. Our studies identified the electrostatic interaction (from free energy calculation) to be the driving stabilising force for the E protein-peptide interactions. Five key E protein residues were also identified that had the most interactions with the peptides: (polar) LYS36, ASN37, and ARG350, and (nonpolar) LEU351 and VAL354; these residues might play crucial roles in the effective binding interactions. One of the peptide fragments, DN58opt_8-13 (PFWFFYRH), showed the best inhibitory activity, at about 63% DENV2 plague reduction, compared with no treatment. This correlates well with the in silico studies in which the peptide possessed the lowest binding energy (-9.0 kcal/mol) and was maintained steadily within the binding pocket of DENV2 E protein during the MD simulations. This study demonstrates the use of computational studies to expand research on lead optimisation of antiviral peptides, thus explaining the inhibitory potential of the designed peptides.
Keywords: dengue; envelope protein; molecular docking; molecular dynamics; peptide; plaque assay.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

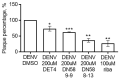

Similar articles
-
Computational-aided design: minimal peptide sequence to block dengue virus transmission into cells.J Biomol Struct Dyn. 2022 Jul;40(11):5026-5035. doi: 10.1080/07391102.2020.1866074. Epub 2020 Dec 31. J Biomol Struct Dyn. 2022. PMID: 33382015
-
Conformational and energy evaluations of novel peptides binding to dengue virus envelope protein.J Mol Graph Model. 2017 Jun;74:273-287. doi: 10.1016/j.jmgm.2017.03.010. Epub 2017 Apr 7. J Mol Graph Model. 2017. PMID: 28458006
-
Molecular docking, simulations of animal peptides against the envelope protein of Dengue virus.J Biomol Struct Dyn. 2024;42(24):13386-13400. doi: 10.1080/07391102.2023.2275183. Epub 2023 Nov 6. J Biomol Struct Dyn. 2024. PMID: 37929876
-
Discovery of Potent Inhibitors for the Inhibition of Dengue Envelope Protein: An In Silico Approach.Curr Top Med Chem. 2018;18(18):1585-1602. doi: 10.2174/1568026618666181025100736. Curr Top Med Chem. 2018. PMID: 30360716 Review.
-
Therapeutic peptides for coronary artery diseases: in silico methods and current perspectives.Amino Acids. 2024 May 31;56(1):37. doi: 10.1007/s00726-024-03397-3. Amino Acids. 2024. PMID: 38822212 Free PMC article. Review.
References
-
- World Health Organization Dengue and Severe Dengue. [(accessed on 1 April 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
-
- Singh A., Robinson A.W.T. Vector Control Interventions to Prevent Dengue: Current Situation and Strategies for Future Improvements to Management of Aedes in India. J. Emerg. Infect. Dis. 2017;2:1000123. doi: 10.4172/2472-4998.1000123. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

