The Evolutionary Dance between Innate Host Antiviral Pathways and SARS-CoV-2
- PMID: 35631059
- PMCID: PMC9147806
- DOI: 10.3390/pathogens11050538
The Evolutionary Dance between Innate Host Antiviral Pathways and SARS-CoV-2
Abstract
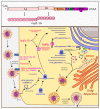
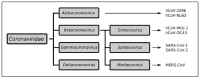
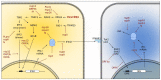
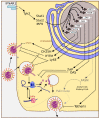
Compared to what we knew at the start of the SARS-CoV-2 global pandemic, our understanding of the interplay between the interferon signaling pathway and SARS-CoV-2 infection has dramatically increased. Innate antiviral strategies range from the direct inhibition of viral components to reprograming the host's own metabolic pathways to block viral infection. SARS-CoV-2 has also evolved to exploit diverse tactics to overcome immune barriers and successfully infect host cells. Herein, we review the current knowledge of the innate immune signaling pathways triggered by SARS-CoV-2 with a focus on the type I interferon response, as well as the mechanisms by which SARS-CoV-2 impairs those defenses.
Keywords: COVID-19; SARS-CoV-2; innate immunity; interferon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Megger D.A., Philipp J., Le-Trilling V.T.K., Sitek B., Trilling M. Deciphering of the Human Interferon-Regulated Proteome by Mass Spectrometry-Based Quantitative Analysis Reveals Extent and Dynamics of Protein Induction and Repression. Front. Immunol. 2017;8:1139. doi: 10.3389/fimmu.2017.01139. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

