Persistent Enterovirus Infection: Little Deletions, Long Infections
- PMID: 35632526
- PMCID: PMC9143164
- DOI: 10.3390/vaccines10050770
Persistent Enterovirus Infection: Little Deletions, Long Infections
Abstract
Enteroviruses have now been shown to persist in cell cultures and in vivo by a novel mechanism involving the deletion of varying amounts of the 5' terminal genomic region termed domain I (also known as the cloverleaf). Molecular clones of coxsackievirus B3 (CVB3) genomes with 5' terminal deletions (TD) of varying length allow the study of these mutant populations, which are able to replicate in the complete absence of wildtype virus genomes. The study of TD enteroviruses has revealed numerous significant differences from canonical enteroviral biology. The deletions appear and become the dominant population when an enterovirus replicates in quiescent cell populations, but can also occur if one of the cis-acting replication elements of the genome (CRE-2C) is artificially mutated in the element's stem and loop structures. This review discusses how the TD genomes arise, how they interact with the host, and their effects on host biology.
Keywords: cis-acting replication element; coxsackievirus B; enterovirus; negative-strand initiation; persistent infection; positive-strand initiation; terminal deletion.
Conflict of interest statement
The author declares no conflict of interest.
Figures
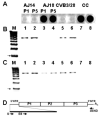

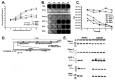
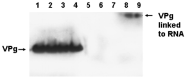


References
-
- Racaniello V.R. Picornaviridae: The Viruses and their Replication. In: Knipe D.M., Howley P.M., Cohen J.I., Griffin D.E., Lamb R.A., Martin M.A., Racaniello V.R., Roizman B., editors. Fields Virology. 6th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 453–489.
-
- Pallansch M.A., Oberste M.S., Whitton J.L. Enteroviruses: Polioviruses, Coxsackieviruses, Echoviruses, and Newer Enteroviruses. In: Knipe D.M., Howley P.M., Cohen J.I., Griffin D.E., Lamb R.A., Martin M.A., Racaniello V.R., Roizman B., editors. Fields Virology. 6th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2001. pp. 490–530.
Publication types
LinkOut - more resources
Full Text Sources

