Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine
- PMID: 35632666
- PMCID: PMC9142977
- DOI: 10.3390/v14050924
Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine
Abstract
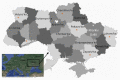
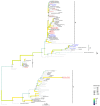
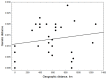
Porcine circovirus type 2 (PCV2) is responsible for a number of porcine circovirus-associated diseases (PCVAD) that can severely impact domestic pig herds. For a non-enveloped virus with a small genome (1.7 kb ssDNA), PCV2 is remarkably diverse, with eight genotypes (a-h). New genotypes of PCV2 can spread through the migration of wild boar, which are thought to infect domestic pigs and spread further through the domestic pig trade. Despite a large swine population, the diversity of PCV2 genotypes in Ukraine has been under-sampled, with few PCV2 genome sequences reported in the past decade. To gain a deeper understanding of PCV2 genotype diversity in Ukraine, samples of blood serum were collected from wild boars (n = 107) that were hunted in Ukraine during the November-December 2012 hunting season. We found 34/107 (31.8%) prevalence of PCV2 by diagnostic PCR. For domestic pigs, liver samples (n = 16) were collected from a commercial market near Kharkiv in 2019, of which 6 out of 16 (37%) samples were positive for PCV2. We sequenced the genotyping locus ORF2, a gene encoding the PCV2 viral capsid (Cap), for 11 wild boar and six domestic pig samples in Ukraine using an Oxford Nanopore MinION device. Of 17 samples with resolved genotypes, the PCV2 genotype b was the most common in wild boar samples (10 out of 11, 91%), while the domestic pigs were infected with genotypes b and d. We also detected genotype b/d and b/a co-infections in wild boars and domestic pigs, respectively, and for the first time in Ukraine we detected genotype f in a wild boar from Poltava. Building a maximum-likelihood phylogeny, we identified a sublineage of PCV2 genotype b infections in both wild and domestic swine, suggesting a possible epizootic cluster and an ecological interaction between wild boar and domestic pig populations in northeastern Ukraine.
Keywords: MinION; PCV2; Ukraine; domestic pig; genotype; phylogenetics; porcine circovirus; wild boar.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- USDA Livestock and Poultry: World Markets and Trade. Global Market Analysis. [(accessed on 1 April 2022)];2021 Available online: https://apps.fas.usda.gov/psdonline/circulars/livestock_poultry.pdf.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

