Multimodal Functionalities of HIV-1 Integrase
- PMID: 35632668
- PMCID: PMC9144474
- DOI: 10.3390/v14050926
Multimodal Functionalities of HIV-1 Integrase
Abstract
Integrase is the retroviral protein responsible for integrating reverse transcripts into cellular genomes. Co-packaged with viral RNA and reverse transcriptase into capsid-encased viral cores, human immunodeficiency virus 1 (HIV-1) integrase has long been implicated in reverse transcription and virion maturation. However, the underlying mechanisms of integrase in these non-catalytic-related viral replication steps have remained elusive. Recent results have shown that integrase binds genomic RNA in virions, and that mutational or pharmacological disruption of integrase-RNA binding yields eccentric virion particles with ribonucleoprotein complexes situated outside of the capsid shell. Such viruses are defective for reverse transcription due to preferential loss of integrase and viral RNA from infected target cells. Parallel research has revealed defective integrase-RNA binding and eccentric particle formation as common features of class II integrase mutant viruses, a phenotypic grouping of viruses that display defects at steps beyond integration. In light of these new findings, we propose three new subclasses of class II mutant viruses (a, b, and c), all of which are defective for integrase-RNA binding and particle morphogenesis, but differ based on distinct underlying mechanisms exhibited by the associated integrase mutant proteins. We also assess how these findings inform the role of integrase in HIV-1 particle maturation.
Keywords: HIV; aberrant integrase multimerization; allosteric integrase inhibitor; integrase; integrase-RNA binding; virus maturation; virus morphogenesis.
Conflict of interest statement
A.N.E. has in the past received fees from ViiV Healthcare, Co. M.K. declares no conflict of interest.
Figures
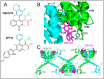


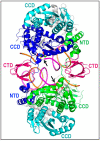
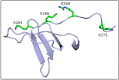

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

