Anomalous HIV-1 RNA, How Cap-Methylation Segregates Viral Transcripts by Form and Function
- PMID: 35632676
- PMCID: PMC9145092
- DOI: 10.3390/v14050935
Anomalous HIV-1 RNA, How Cap-Methylation Segregates Viral Transcripts by Form and Function
Abstract
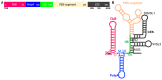
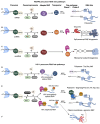
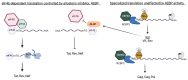
The acquisition of m7G-cap-binding proteins is now recognized as a major variable driving the form and function of host RNAs. This manuscript compares the 5'-cap-RNA binding proteins that engage HIV-1 precursor RNAs, host mRNAs, small nuclear (sn)- and small nucleolar (sno) RNAs and sort into disparate RNA-fate pathways. Before completion of the transcription cycle, the transcription start site of nascent class II RNAs is appended to a non-templated guanosine that is methylated (m7G-cap) and bound by hetero-dimeric CBP80-CBP20 cap binding complex (CBC). The CBC is a nexus for the co-transcriptional processing of precursor RNAs to mRNAs and the snRNA and snoRNA of spliceosomal and ribosomal ribonucleoproteins (RNPs). Just as sn/sno-RNAs experience hyper-methylation of m7G-cap to trimethylguanosine (TMG)-cap, so do select HIV RNAs and an emerging cohort of mRNAs. TMG-cap is blocked from Watson:Crick base pairing and disqualified from participating in secondary structure. The HIV TMG-cap has been shown to license select viral transcripts for specialized cap-dependent translation initiation without eIF4E that is dependent upon CBP80/NCBP3. The exceptional activity of HIV precursor RNAs secures their access to maturation pathways of sn/snoRNAs, canonical and non-canonical host mRNAs in proper stoichiometry to execute the retroviral replication cycle.
Keywords: NCBP3; RNA virus; epigenetic modification; internal ribosome entry; junD; ribosome scanning; specialized translation; trimethylguanosine (TMG) cap.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design or the writing of the manuscript.
Figures
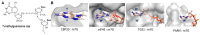


References
-
- Coffin J.M., Hughes S.H., Varmus H. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY, USA: 1997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

