Duplex One-Step RT-qPCR Assays for Simultaneous Detection of Genomic and Subgenomic RNAs of SARS-CoV-2 Variants
- PMID: 35632807
- PMCID: PMC9143037
- DOI: 10.3390/v14051066
Duplex One-Step RT-qPCR Assays for Simultaneous Detection of Genomic and Subgenomic RNAs of SARS-CoV-2 Variants
Abstract
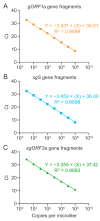
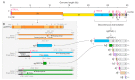
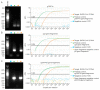
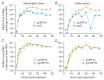
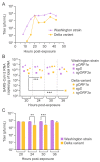
A hallmark of severe acute respiratory syndrome virus (SARS-CoV-2) replication is the discontinuous transcription of open reading frames (ORFs) encoding structural virus proteins. Real-time reverse transcription PCR (RT-qPCR) assays in previous publications used either single or multiplex assays for SARS-CoV-2 genomic RNA detection and a singleplex approach for subgenomic RNA detection. Although multiplex approaches often target multiple genomic RNA segments, an assay that concurrently detects genomic and subgenomic targets has been lacking. To bridge this gap, we developed two duplex one-step RT-qPCR assays that detect SARS-CoV-2 genomic ORF1a and either subgenomic spike or subgenomic ORF3a RNAs. All primers and probes for our assays were designed to bind to variants of SARS-CoV-2. In this study, our assays successfully detected SARS-CoV-2 Washington strain and delta variant isolates at various time points during the course of live virus infection in vitro. The ability to quantify subgenomic SARS-CoV-2 RNA is important, as it may indicate the presence of active replication, particularly in samples collected longitudinally. Furthermore, specific detection of genomic and subgenomic RNAs simultaneously in a single reaction increases assay efficiency, potentially leading to expedited lucidity about viral replication and pathogenesis of any variant of SARS-CoV-2.
Keywords: RNA; SARS-CoV-2; duplex RT-qPCR; genomic; subgenomic; variant.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Figures

Similar articles
-
Profiling of SARS-CoV-2 Subgenomic RNAs in Clinical Specimens.Microbiol Spectr. 2022 Apr 27;10(2):e0018222. doi: 10.1128/spectrum.00182-22. Epub 2022 Mar 21. Microbiol Spectr. 2022. PMID: 35311586 Free PMC article.
-
Characterizing SARS-CoV-2 Transcription of Subgenomic and Genomic RNAs During Early Human Infection Using Multiplexed Droplet Digital Polymerase Chain Reaction.J Infect Dis. 2023 Apr 18;227(8):981-992. doi: 10.1093/infdis/jiac472. J Infect Dis. 2023. PMID: 36468309 Free PMC article.
-
Novel RT-ddPCR assays for measuring the levels of subgenomic and genomic SARS-CoV-2 transcripts.Methods. 2022 May;201:15-25. doi: 10.1016/j.ymeth.2021.04.011. Epub 2021 Apr 18. Methods. 2022. PMID: 33882362 Free PMC article.
-
Subgenomic RNAs and Their Encoded Proteins Contribute to the Rapid Duplication of SARS-CoV-2 and COVID-19 Progression.Biomolecules. 2022 Nov 12;12(11):1680. doi: 10.3390/biom12111680. Biomolecules. 2022. PMID: 36421694 Free PMC article. Review.
-
SARS-CoV-2 Subgenomic RNAs: Characterization, Utility, and Perspectives.Viruses. 2021 Sep 24;13(10):1923. doi: 10.3390/v13101923. Viruses. 2021. PMID: 34696353 Free PMC article. Review.
Cited by
-
Abortive infection of bat fibroblasts with SARS-CoV-2.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2406773121. doi: 10.1073/pnas.2406773121. Epub 2024 Oct 14. Proc Natl Acad Sci U S A. 2024. PMID: 39401365 Free PMC article.
-
Variations in the persistence of 5'-end genomic and subgenomic SARS-CoV-2 RNAs in wastewater from aircraft, airports and wastewater treatment plants.Heliyon. 2024 Apr 16;10(9):e29703. doi: 10.1016/j.heliyon.2024.e29703. eCollection 2024 May 15. Heliyon. 2024. PMID: 38694057 Free PMC article.
References
-
- Boshier F.A.T., Pang J., Penner J., Parker M., Alders N., Bamford A., Grandjean L., Grunewald S., Hatcher J., Best T., et al. Evolution of viral variants in remdesivir-treated and untreated SARS-CoV-2-infected pediatrics patients. J. Med. Virol. 2022;94:161–172. doi: 10.1002/jmv.27285. - DOI - PMC - PubMed
-
- Johns Hopkins University & Medicine Coronavirus Resource Center COVID-19 Dashboard. 2022. [(accessed on 4 January 2022)]. Available online: https://coronavirus.jhu.edu/map.html.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

