Bridging the gap between prostate radiology and pathology through machine learning
- PMID: 35633505
- PMCID: PMC9543295
- DOI: 10.1002/mp.15777
Bridging the gap between prostate radiology and pathology through machine learning
Abstract
Background: Prostate cancer remains the second deadliest cancer for American men despite clinical advancements. Currently, magnetic resonance imaging (MRI) is considered the most sensitive non-invasive imaging modality that enables visualization, detection, and localization of prostate cancer, and is increasingly used to guide targeted biopsies for prostate cancer diagnosis. However, its utility remains limited due to high rates of false positives and false negatives as well as low inter-reader agreements.
Purpose: Machine learning methods to detect and localize cancer on prostate MRI can help standardize radiologist interpretations. However, existing machine learning methods vary not only in model architecture, but also in the ground truth labeling strategies used for model training. We compare different labeling strategies and the effects they have on the performance of different machine learning models for prostate cancer detection on MRI.
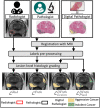
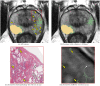
Methods: Four different deep learning models (SPCNet, U-Net, branched U-Net, and DeepLabv3+) were trained to detect prostate cancer on MRI using 75 patients with radical prostatectomy, and evaluated using 40 patients with radical prostatectomy and 275 patients with targeted biopsy. Each deep learning model was trained with four different label types: pathology-confirmed radiologist labels, pathologist labels on whole-mount histopathology images, and lesion-level and pixel-level digital pathologist labels (previously validated deep learning algorithm on histopathology images to predict pixel-level Gleason patterns) on whole-mount histopathology images. The pathologist and digital pathologist labels (collectively referred to as pathology labels) were mapped onto pre-operative MRI using an automated MRI-histopathology registration platform.
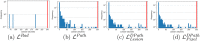
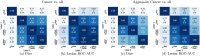
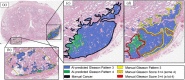
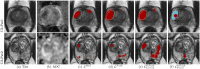
Results: Radiologist labels missed cancers (ROC-AUC: 0.75-0.84), had lower lesion volumes (~68% of pathology lesions), and lower Dice overlaps (0.24-0.28) when compared with pathology labels. Consequently, machine learning models trained with radiologist labels also showed inferior performance compared to models trained with pathology labels. Digital pathologist labels showed high concordance with pathologist labels of cancer (lesion ROC-AUC: 0.97-1, lesion Dice: 0.75-0.93). Machine learning models trained with digital pathologist labels had the highest lesion detection rates in the radical prostatectomy cohort (aggressive lesion ROC-AUC: 0.91-0.94), and had generalizable and comparable performance to pathologist label-trained-models in the targeted biopsy cohort (aggressive lesion ROC-AUC: 0.87-0.88), irrespective of the deep learning architecture. Moreover, machine learning models trained with pixel-level digital pathologist labels were able to selectively identify aggressive and indolent cancer components in mixed lesions on MRI, which is not possible with any human-annotated label type.
Conclusions: Machine learning models for prostate MRI interpretation that are trained with digital pathologist labels showed higher or comparable performance with pathologist label-trained models in both radical prostatectomy and targeted biopsy cohort. Digital pathologist labels can reduce challenges associated with human annotations, including labor, time, inter- and intra-reader variability, and can help bridge the gap between prostate radiology and pathology by enabling the training of reliable machine learning models to detect and localize prostate cancer on MRI.
Keywords: aggressive versus indolent cancer; cancer labels; deep learning; digital pathology; prostate MRI.
© 2022 The Authors. Medical Physics published by Wiley Periodicals LLC on behalf of American Association of Physicists in Medicine.
Conflict of interest statement
Mirabela Rusu has research grants from GE Healthcare and Philips Healthcare.
Figures




References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7‐33. - PubMed
-
- Ahmed HU, Bosaily AE‐S, Brown LC, et al. Diagnostic accuracy of multi‐parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815‐822. - PubMed
-
- Johnson DC, Raman SS, Mirak SA, et al. Detection of individual prostate cancer foci via multiparametric magnetic resonance imaging. Eur Urol. 2019;75:712‐720. - PubMed
-
- van der Leest M, Cornel E, Israel B, et al. Head‐to‐head comparison of transrectal ultrasound‐guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance‐guided biopsy in biopsy‐naïve men with elevated prostate‐specific antigen: a large prospective multicenter clinical study. Eur Urol. 2019;75:570‐578. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

