Molecular Typing and Rapid Identification of Human Adenoviruses Associated With Respiratory Diseases Using Universal PCR and Sequencing Primers for the Three Major Capsid Genes: Penton Base, Hexon, and Fiber
- PMID: 35633710
- PMCID: PMC9133664
- DOI: 10.3389/fmicb.2022.911694
Molecular Typing and Rapid Identification of Human Adenoviruses Associated With Respiratory Diseases Using Universal PCR and Sequencing Primers for the Three Major Capsid Genes: Penton Base, Hexon, and Fiber
Abstract
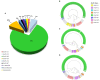
Human adenoviruses (HAdVs) within species B, C, and E are responsible for highly contagious and potentially severe respiratory disease infections. The traditional method to type these pathogens was based on virus neutralization and hemagglutination assays, which are both time-consuming and difficult, particularly due to the nonavailability of reagents. Subsequent molecular typing based on the partial characterization of the hexon gene and/or the restriction enzyme analysis (REA) of the genomes is inadequate, particularly in identifying recombinants. Here, a rapid, simple, and cost-effective method for molecular typing HAdV respiratory pathogens is presented. This incorporates three pairs of universal PCR primers that target the variable regions of the three major capsid genes, i.e., hexon, penton base, and fiber genes, that span the genome. The protocol enables typing and characterization of genotypes within species B, C, and E, as well as of some genotypes within species D and F. To validate this method, we surveyed 100 children with HAdV-associated acute respiratory infections identified by direct immunofluorescence (Hong Kong; July through October, 2014). Throat swab specimens were collected and analyzed by PCR amplification and sequencing; these sequences were characterized by BLAST. HAdVs were detected in 98 out of 100 (98%) samples, distributing as follows: 74 HAdV-B3 (74%); 10 HAdV-E4 (10%); 7 HAdV-C2 (7%); 2 HAdV-C6 (2%); 1 HAdV-B7 (1%); 1 HAdV-C1 (1%); 2 co-infection (2%); and 1 novel recombinant (1%). This study is the first detailed molecular epidemiological survey of HAdVs in Hong Kong. The developed method allows for the rapid identification of HAdV respiratory pathogens, including recombinants, and bypasses the need for whole genome sequencing for real-time surveillance of circulating adenovirus strains in outbreaks and populations by clinical virologists, public health officials, and epidemiologists.
Keywords: Hong Kong; adenovirus; co-infection; epidemiology; molecular typing; recombination; universal primers.
Copyright © 2022 Wu, Zhang, Lan, Quan, Ou, Zhao, Wu, Woo, Seto and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Chen Y., Liu F., Wang C., Zhao M., Deng L., Zhong J., et al. (2016). Molecular identification and epidemiological features of human adenoviruses associated with acute respiratory infections in hospitalized children in southern China, 2012-2013. PLoS One 11:e0155412. doi: 10.1371/journal.pone.0155412, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

