Expression, Purification, and In Silico Characterization of Mycobacterium smegmatis Alternative Sigma Factor SigB
- PMID: 35634445
- PMCID: PMC9142298
- DOI: 10.1155/2022/7475704
Expression, Purification, and In Silico Characterization of Mycobacterium smegmatis Alternative Sigma Factor SigB
Abstract
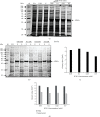
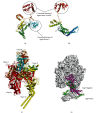
Sigma factor B (SigB), an alternative sigma factor (ASF), is very similar to primary sigma factor SigA (σ 70) but dispensable for growth in both Mycobacterium smegmatis (Msmeg) and Mycobacterium tuberculosis (Mtb). It is involved in general stress responses including heat, oxidative, surface, starvation stress, and macrophage infections. Despite having an extremely short half-life, SigB tends to operate downstream of at least three stress-responsive extra cytoplasmic function (ECF) sigma factors (SigH, SigE, SigL) and SigF involved in multiple signaling pathways. There is very little information available regarding the regulation of SigB sigma factor and its interacting protein partners. Hence, we cloned the SigB gene into pET28a vector and optimized its expression in three different strains of E. coli, viz., (BL21 (DE3), C41 (DE3), and CodonPlus (DE3)). We also optimized several other parameters for the expression of recombinant SigB including IPTG concentration, temperature, and time duration. We achieved the maximum expression of SigB at 25°C in the soluble fraction of the cell which was purified by affinity chromatography using Ni-NTA and further confirmed by Western blotting. Further, structural characterization demonstrates the instability of SigB in comparison to SigA that is carried out using homology modeling and structure function relationship. We have done protein-protein docking of RNA polymerase (RNAP) of Msmeg and SigB. This effort provides a platform for pulldown assay, structural, and other studies with the recombinant protein to deduce the SigB interacting proteins, which might pave the way to study its signaling networks along with its regulation.
Copyright © 2022 Rakesh Kumar Singh et al.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

