Precision health diagnostic and surveillance network uses S gene target failure (SGTF) combined with sequencing technologies to track emerging SARS-CoV-2 variants
- PMID: 35634961
- PMCID: PMC9092005
- DOI: 10.1002/iid3.634
Precision health diagnostic and surveillance network uses S gene target failure (SGTF) combined with sequencing technologies to track emerging SARS-CoV-2 variants
Abstract
Introduction: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic revealed a worldwide lack of effective molecular surveillance networks at local, state, and national levels, which are essential to identify, monitor, and limit viral community spread. SARS-CoV-2 variants of concern (VOCs) such as Alpha and Omicron, which show increased transmissibility and immune evasion, rapidly became dominant VOCs worldwide. Our objective was to develop an evidenced-based genomic surveillance algorithm, combining reverse transcription polymerase chain reaction (RT-PCR) and sequencing technologies to quickly identify highly contagious VOCs, before cases accumulate exponentially.
Methods: Deidentified data were obtained from 508,969 patients tested for coronavirus disease 2019 (COVID-19) with the TaqPath COVID-19 RT-PCR Combo Kit (ThermoFisher) in four CLIA-certified clinical laboratories in Puerto Rico (n = 86,639) and in three CLIA-certified clinical laboratories in the United States (n = 422,330).
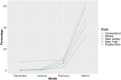
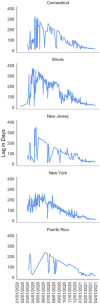
Results: TaqPath data revealed a frequency of S Gene Target Failure (SGTF) > 47% for the last week of March 2021 in both, Puerto Rico and US laboratories. The monthly frequency of SGTF in Puerto Rico steadily increased exponentially from 4% in November 2020 to 47% in March 2021. The weekly SGTF rate in US samples was high (>8%) from late December to early January and then also increased exponentially through April (48%). The exponential increase in SGFT prevalence in Puerto Rico was concurrent with a sharp increase in VOCs among all SARS-CoV-2 sequences from Puerto Rico uploaded to Global Influenza Surveillance and Response System (GISAID) (n = 461). Alpha variant frequency increased from <1% in the last week of January 2021 to 51.5% of viral sequences from Puerto Rico collected in the last week of March 2021.
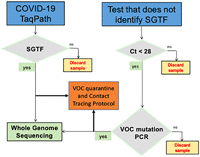
Conclusions: According to the proposed evidence-based algorithm, approximately 50% of all SGTF patients should be managed with VOCs self-quarantine and contact tracing protocols, while WGS confirms their lineage in genomic surveillance laboratories. Our results suggest this workflow is useful for tracking VOCs with SGTF.
Keywords: SARS-CoV-2; algorithms; genomics; population surveillance; precision medicine; public health.
© 2022 The Authors. Immunity, Inflammation and Disease published by John Wiley & Sons Ltd.
Conflict of interest statement
Matthew J. MacKay, Rachel Baits, Daisy Salgado, Gaurav Khullar, Jessica Metti, Timothy Baker, Joel Dudley, and Christopher E. Mason, work for Tempus Labs. Rafael Guerrero‐Preston works for LifeGene‐Biomarks. The remaining authors declare no conflict of interest.
Figures

References
-
- Rabaan AA, Al‐Ahmed SH, Haque S, et al. SARS‐CoV‐2, SARS‐CoV, and MERS‐COV: a comparative overview. Infez Med. 2020;28(2):174‐184. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

