Epidemiological and genomic findings of the first documented Italian outbreak of SARS-CoV-2 Alpha variant of concern
- PMID: 35636310
- PMCID: PMC9098518
- DOI: 10.1016/j.epidem.2022.100578
Epidemiological and genomic findings of the first documented Italian outbreak of SARS-CoV-2 Alpha variant of concern
Abstract
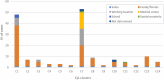
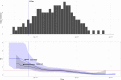
From 24 December 2020 to 8 February 2021, 163 cases of SARS-CoV-2 Alpha variant of concern (VOC) were identified in Chieti province, Abruzzo region. Epidemiological data allowed the identification of 14 epi-clusters. With one exception, all the epi-clusters were linked to the town of Guardiagrele: 149 contacts formed the network, two-thirds of which were referred to the family/friends context. Real data were then used to estimate transmission parameters. According to our method, the calculated Re(t) was higher than 2 before the 12 December 2020. Similar values were obtained from other studies considering Alpha VOC. Italian sequence data were combined with a random subset of sequences obtained from the GISAID database. Genomic analysis showed close identity between the sequences from Guardiagrele, forming one distinct clade. This would suggest one or limited unspecified viral introductions from outside to Abruzzo region in early December 2020, which led to the diffusion of Alpha VOC in Guardiagrele and in neighbouring municipalities, with very limited inter-regional mixing.
Keywords: Abruzzo; Epidemiology; Genomic surveillance; Reproduction number; SARS-CoV-2, Alpha variant.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Abbott, S., 2020. epiforecasts/EpiNow2: Initial release [WWW Document]. 10.5281/ZENODO.3957490. - DOI
-
- Abbott S., Hellewell J., Sherratt K., Gostic K., Hickson J., Badr H.S., DeWitt M., Thompson R., EpiForecasts, Funk S. (2020a). EpiNow2: Estimate Real-Time Case Counts and Time-Varying Epidemiological Parameters. doi: 10.5281/zenodo.3957489.
-
- Abbott S., Hellewell J., Thompson R.N., Sherratt K., Gibbs H.P., Bosse N.I., Munday J.D., Meakin S., Doughty E.L., Chun J.Y., Chan Y.-W.D., Finger F., Campbell P., Endo A., Pearson C.A.B., Gimma A., Russell T., Flasche S., Kucharski A.J., Eggo R.M., Funk S. Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts. Wellcome Open Res. 2020;5:112. doi: 10.12688/wellcomeopenres.16006.2. - DOI
-
- Amato L., Jurisic L., Puglia I., Di Lollo V., Curini V., Torzi G., Di Girolamo A., Mangone I., Mancinelli A., Decaro N., Calistri P., Di Giallonardo F., Lorusso A., D’Alterio N. Multiple detection and spread of novel strains of the SARS-CoV-2 B.1.177 (B.1.177.75) lineage that test negative by a commercially available nucleocapsid gene real-time RT-PCR. Emerging Microbes & Infections. 2021:1148–1155. doi: 10.1080/22221751.2021.1933609. - DOI - PMC - PubMed
-
- Bager P., Wohlfahrt J., Fonager J., Rasmussen M., Albertsen M., Michaelsen T.Y., Møller C.H., Ethelberg S., Legarth R., Button M.S.F., Gubbels S., Voldstedlund M., Mølbak K., Skov R.L., Fomsgaard A., Krause T.G. Risk of hospitalisation associated with infection with SARS-CoV-2 lineage B.1.1.7 in Denmark: an observational cohort study. Lancet Infect. Dis. 2021 doi: 10.1016/s1473-3099(21)00290-5. - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

