A lncRNA signature associated with tumor immune heterogeneity predicts distant metastasis in locoregionally advanced nasopharyngeal carcinoma
- PMID: 35637194
- PMCID: PMC9151760
- DOI: 10.1038/s41467-022-30709-6
A lncRNA signature associated with tumor immune heterogeneity predicts distant metastasis in locoregionally advanced nasopharyngeal carcinoma
Abstract
Increasing evidence has revealed the roles of long noncoding RNAs (lncRNAs) as tumor biomarkers. Here, we introduce an immune-associated nine-lncRNA signature for predicting distant metastasis in locoregionally advanced nasopharyngeal carcinoma (LA-NPC). The nine lncRNAs are identified through microarray profiling, followed by RT-qPCR validation and selection using a machine learning method in the training cohort (n = 177). This nine-lncRNA signature classifies patients into high and low risk groups, which have significantly different distant metastasis-free survival. Validations in the Guangzhou internal (n = 177) and Guilin external (n = 150) cohorts yield similar results, confirming that the signature is an independent risk factor for distant metastasis and outperforms anatomy-based metrics in identifying patients with high metastatic risk. Integrative analyses show that this nine-lncRNA signature correlates with immune activity and lymphocyte infiltration, which is validated by digital pathology. Our results suggest that the immune-associated nine-lncRNA signature can serve as a promising biomarker for metastasis prediction in LA-NPC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
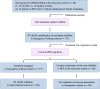
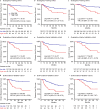

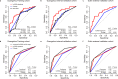
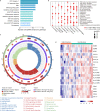
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

