Carotenoid binding in Gloeobacteria rhodopsin provides insights into divergent evolution of xanthorhodopsin types
- PMID: 35637261
- PMCID: PMC9151804
- DOI: 10.1038/s42003-022-03429-2
Carotenoid binding in Gloeobacteria rhodopsin provides insights into divergent evolution of xanthorhodopsin types
Abstract
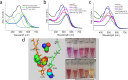
The position of carotenoid in xanthorhodopsin has been elucidated. However, a challenging expression of this opsin and a complex biosynthesis carotenoid in the laboratory hold back the insightful study of this rhodopsin. Here, we demonstrated co-expression of the xanthorhodopsin type isolated from Gloeobacter violaceus PCC 7421-Gloeobacter rhodopsin (GR) with a biosynthesized keto-carotenoid (canthaxanthin) targeting the carotenoid binding site. Direct mutation-induced changes in carotenoid-rhodopsin interaction revealed three crucial features: (1) carotenoid locked motif (CLM), (2) carotenoid aligned motif (CAM), and color tuning serines (CTS). Our single mutation results at 178 position (G178W) confirmed inhibition of carotenoid binding; however, the mutants showed better stability and proton pumping, which was also observed in the case of carotenoid binding characteristics. These effects demonstrated an adaptation of microbial rhodopsin that diverges from carotenoid harboring, along with expression in the dinoflagellate Pyrocystis lunula rhodopsin and the evolutionary substitution model. The study highlights a critical position of the carotenoid binding site, which significantly allows another protein engineering approach in the microbial rhodopsin family.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

