CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis
- PMID: 35637331
- PMCID: PMC9159947
- DOI: 10.1038/s41564-022-01130-y
CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis
Abstract
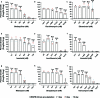
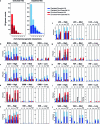
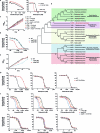
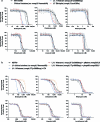
Mycobacterium tuberculosis (Mtb) infection is notoriously difficult to treat. Treatment efficacy is limited by Mtb's intrinsic drug resistance, as well as its ability to evolve acquired resistance to all antituberculars in clinical use. A deeper understanding of the bacterial pathways that influence drug efficacy could facilitate the development of more effective therapies, identify new mechanisms of acquired resistance, and reveal overlooked therapeutic opportunities. Here we developed a CRISPR interference chemical-genetics platform to titrate the expression of Mtb genes and quantify bacterial fitness in the presence of different drugs. We discovered diverse mechanisms of intrinsic drug resistance, unveiling hundreds of potential targets for synergistic drug combinations. Combining chemical genetics with comparative genomics of Mtb clinical isolates, we further identified several previously unknown mechanisms of acquired drug resistance, one of which is associated with a multidrug-resistant tuberculosis outbreak in South America. Lastly, we found that the intrinsic resistance factor whiB7 was inactivated in an entire Mtb sublineage endemic to Southeast Asia, presenting an opportunity to potentially repurpose the macrolide antibiotic clarithromycin to treat tuberculosis. This chemical-genetic map provides a rich resource to understand drug efficacy in Mtb and guide future tuberculosis drug development and treatment.
© 2022. The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures












Comment in
-
Gene-drug potency screening in M. tuberculosis.Nat Microbiol. 2022 Jun;7(6):743-744. doi: 10.1038/s41564-022-01139-3. Nat Microbiol. 2022. PMID: 35637332 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

