Ribosome stalling during selenoprotein translation exposes a ferroptosis vulnerability
- PMID: 35637349
- PMCID: PMC9469796
- DOI: 10.1038/s41589-022-01033-3
Ribosome stalling during selenoprotein translation exposes a ferroptosis vulnerability
Abstract
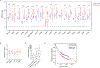
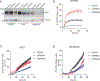
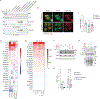
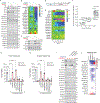
The selenoprotein glutathione peroxidase 4 (GPX4) prevents ferroptosis by converting lipid peroxides into nontoxic lipid alcohols. GPX4 has emerged as a promising therapeutic target for cancer treatment, but some cancer cells are resistant to ferroptosis triggered by GPX4 inhibition. Using a chemical-genetic screen, we identify LRP8 (also known as ApoER2) as a ferroptosis resistance factor that is upregulated in cancer. Loss of LRP8 decreases cellular selenium levels and the expression of a subset of selenoproteins. Counter to the canonical hierarchical selenoprotein regulatory program, GPX4 levels are strongly reduced due to impaired translation. Mechanistically, low selenium levels result in ribosome stalling at the inefficiently decoded GPX4 selenocysteine UGA codon, leading to ribosome collisions, early translation termination and proteasomal clearance of the N-terminal GPX4 fragment. These findings reveal rewiring of the selenoprotein hierarchy in cancer cells and identify ribosome stalling and collisions during GPX4 translation as ferroptosis vulnerabilities in cancer.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS
S.J.D. is a co-founder of Prothegen Inc. and a member of the advisory board for Ferro Therapeutics and Hillstream Biopharma. S.J.D. and J.A.O. have patent applications related to ferroptosis.
Figures












Comment in
-
Loss of LPR8 Reduces GPX4 Levels, Resulting in Ferroptosis Sensitivity.Cancer Discov. 2022 Aug 5;12(8):1835. doi: 10.1158/2159-8290.CD-RW2022-109. Cancer Discov. 2022. PMID: 35713354
References
-
- Doll S et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

