Individual variability in brain representations of pain
- PMID: 35637368
- PMCID: PMC9435464
- DOI: 10.1038/s41593-022-01081-x
Individual variability in brain representations of pain
Abstract
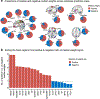
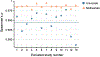
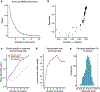
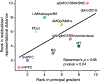
Characterizing cerebral contributions to individual variability in pain processing is crucial for personalized pain medicine, but has yet to be done. In the present study, we address this problem by identifying brain regions with high versus low interindividual variability in their relationship with pain. We trained idiographic pain-predictive models with 13 single-trial functional MRI datasets (n = 404, discovery set) and quantified voxel-level importance for individualized pain prediction. With 21 regions identified as important pain predictors, we examined the interindividual variability of local pain-predictive weights in these regions. Higher-order transmodal regions, such as ventromedial and ventrolateral prefrontal cortices, showed larger individual variability, whereas unimodal regions, such as somatomotor cortices, showed more stable pain representations across individuals. We replicated this result in an independent dataset (n = 124). Overall, our study identifies cerebral sources of individual differences in pain processing, providing potential targets for personalized assessment and treatment of pain.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures