A local human Vδ1 T cell population is associated with survival in nonsmall-cell lung cancer
- PMID: 35637401
- PMCID: PMC9236901
- DOI: 10.1038/s43018-022-00376-z
A local human Vδ1 T cell population is associated with survival in nonsmall-cell lung cancer
Abstract
Murine tissues harbor signature γδ T cell compartments with profound yet differential impacts on carcinogenesis. Conversely, human tissue-resident γδ cells are less well defined. In the present study, we show that human lung tissues harbor a resident Vδ1 γδ T cell population. Moreover, we demonstrate that Vδ1 T cells with resident memory and effector memory phenotypes were enriched in lung tumors compared with nontumor lung tissues. Intratumoral Vδ1 T cells possessed stem-like features and were skewed toward cytolysis and helper T cell type 1 function, akin to intratumoral natural killer and CD8+ T cells considered beneficial to the patient. Indeed, ongoing remission post-surgery was significantly associated with the numbers of CD45RA-CD27- effector memory Vδ1 T cells in tumors and, most strikingly, with the numbers of CD103+ tissue-resident Vδ1 T cells in nonmalignant lung tissues. Our findings offer basic insights into human body surface immunology that collectively support integrating Vδ1 T cell biology into immunotherapeutic strategies for nonsmall cell lung cancer.
© 2022. The Author(s).
Conflict of interest statement
D.B. has consulted for NanoString, reports honoraria from AstraZeneca and has a patent (PCT/GB2020/050221) issued on methods for cancer prognostication. J.R. and M.A.B. have consulted for Achilles Therapeutics. N.M. has stock options in and has consulted for Achilles Therapeutics. N.M. holds European patents relating to targeting neoantigens (PCT/EP2016/059401), identifying patient response to immune checkpoint blockade (PCT/EP2016/071471), determining HLA loss of heterozygosity (PCT/GB2018/052004) and predicting survival rates of patients with cancer (PCT/GB2020/050221). A.H. attended one advisory board for Abbvie, Roche and GRAIL, and reports personal fees from Abbvie, Boehringer Ingelheim, Takeda, AstraZeneca, Daiichi Sankyo, Merck Serono, Merck/MSD, UCB and Roche for delivering general education/training in clinical trials. A.H. owned shares in Illumina and Thermo Fisher Scientific (sold in 2020) and receives fees for membership of Independent Data Monitoring Committees for Roche-sponsored clinical trials. S.A.Q. is co-founder and Chief Scientific Officer of Achilles Therapeutics. A.C.H. is a board member and equity holder in ImmunoQure, AG and Gamma Delta Therapeutics, and is an equity holder in Adaptate Biotherapeutics and chair of the scientific advisory board. C.S. acknowledges grant support from Pfizer, AstraZeneca, Bristol Myers Squibb, Roche-Ventana, Boehringer Ingelheim, Archer Dx Inc (collaboration in minimal residual disease-sequencing technologies) and Ono Pharmaceuticals, is an AstraZeneca Advisory Board member and Chief Investigator for the MeRmaiD1 clinical trial. C.S has consulted for Amgen, AstraZeneca, Bicycle Therapeutics, Bristol Myers Squibb, Celgene, Genentech, GlaxoSmithKline, GRAIL, Illumina, Medixci, Metabomed, MSD, Novartis, Pfizer, Roche-Ventana and Sarah Cannon Research Institute. C.S. has stock options in Apogen Biotechnologies, Epic Biosciences and GRAIL, and has stock options and is co-founder of Achilles Therapeutics. C.S. holds patents relating: to assay technology to detect tumor recurrence (PCT/GB2017/053289); to targeting neoantigens (PCT/EP2016/059401), identifying patent response to immune checkpoint blockade (PCT/EP2016/071471), determining HLA loss of heterozygosity (PCT/GB2018/052004), predicting survival rates of patients with cancer (PCT/GB2020/050221); to treating cancer by targeting Insertion/deletion (indel) mutations (PCT/GB2018/051893); to identifying indel mutation targets (PCT/GB2018/051892); to methods for lung cancer detection (PCT/US2017/028013); and to identifying responders to cancer treatment (PCT/GB2018/051912). The remaining authors declare no competing interests.
Figures
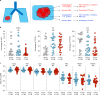


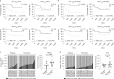


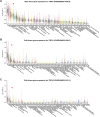

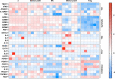

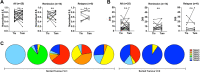
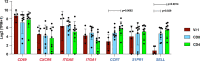
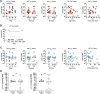
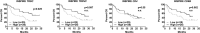
Comment in
-
γδ T cells share the spotlight in cancer.Nat Cancer. 2022 Jun;3(6):657-658. doi: 10.1038/s43018-022-00396-9. Nat Cancer. 2022. PMID: 35764741 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- 21999/CRUK_/Cancer Research UK/United Kingdom
- 30025/CRUK_/Cancer Research UK/United Kingdom
- FC001169 /WT_/Wellcome Trust/United Kingdom
- 22246/CRUK_/Cancer Research UK/United Kingdom
- FC001169/WT_/Wellcome Trust/United Kingdom
- 220589/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- MR/V033077/1/MRC_/Medical Research Council/United Kingdom
- C11496/A30025/CRUK_/Cancer Research UK/United Kingdom
- 29569/CRUK_/Cancer Research UK/United Kingdom
- FC001169 /CRUK_/Cancer Research UK/United Kingdom
- FC001169 /MRC_/Medical Research Council/United Kingdom
- MC_PC_17179/MRC_/Medical Research Council/United Kingdom
- 17786/CRUK_/Cancer Research UK/United Kingdom
- 203141/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- DH_/Department of Health/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials

