Integrated Bioinformatics and Clinical Correlation Analysis of Key Genes, Pathways, and Potential Therapeutic Agents Related to Diabetic Nephropathy
- PMID: 35637650
- PMCID: PMC9148260
- DOI: 10.1155/2022/9204201
Integrated Bioinformatics and Clinical Correlation Analysis of Key Genes, Pathways, and Potential Therapeutic Agents Related to Diabetic Nephropathy
Abstract
Background: Diabetic nephropathy (DN) is a common microvascular complication of diabetes and a major cause of end-stage renal disease, resulting in a substantial socioeconomic burden around the world. Some unknown biomarkers, mechanisms, and potential novel agents regarding DN are yet to be identified.
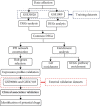
Methods: GSE30528 and GSE1009 were downloaded as training datasets to identify differentially expressed genes (DEGs) of DN. Common DEGs were selected for further analysis. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of DEGs were performed to explore molecular mechanisms and pathways. Protein-protein interaction (PPI) network of DEGs was used to identify the top 10 hub genes of DN. Expression profiles of the hub genes were validated in GSE96804 and GSE47183 datasets. The clinical correlation analyses were conducted to confirm the association between key genes and clinical characteristics in the Nephroseq v5 database. The Drug Gene Interaction Database was used to predict potential targeted drugs.
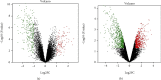
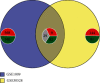
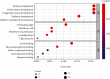
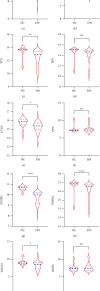
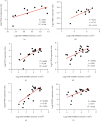
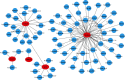
Results: 345 and 1228 DEGs were identified in GSE30528 and GSE1009, respectively; and 120 common DEGs were found. The biological process of DEGs was significantly enriched in kidney development. PI3K-Akt signaling pathway, focal adhesion, complement and coagulation cascades were significantly enriched KEGG pathways. The identified top10 hub genes were VEGFA, NPHS1, WT1, TJP1, CTGF, FYN, SYNPO, PODXL, TNNT2, and BMP2. VEGFA, NPHS1, WT1, CTGF, SYNPO, PODXL, and TNNT2 were significantly downregulated in DN. VEGFA, NPHS1, WT1, CTGF, SYNPO, and PODXL were positively correlated with glomerular filtration rate. The targeted drugs or molecular compounds were enalapril, sildenafil, and fenofibrate target for VEGFA; losartan target for NPHS1; halofuginone, deferoxamine, curcumin, and sirolimus target for WT1; and purpurogallin target for TNNT2.
Conclusions: VEGFA, NPHS1, WT1, CTGF, SYNPO, and PODXL are promising biomarkers for diagnosing and evaluating the progression of DN. The drug-gene interaction analyses provide a list of candidate drugs for the precise treatment of DN.
Copyright © 2022 Shengnan Chen et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

