Adverse effects following anti-COVID-19 vaccination with mRNA-based BNT162b2 are alleviated by altering the route of administration and correlate with baseline enrichment of T and NK cell genes
- PMID: 35639676
- PMCID: PMC9154185
- DOI: 10.1371/journal.pbio.3001643
Adverse effects following anti-COVID-19 vaccination with mRNA-based BNT162b2 are alleviated by altering the route of administration and correlate with baseline enrichment of T and NK cell genes
Abstract
Ensuring high vaccination and even booster vaccination coverage is critical in preventing severe Coronavirus Disease 2019 (COVID-19). Among the various COVID-19 vaccines currently in use, the mRNA vaccines have shown remarkable effectiveness. However, systemic adverse events (AEs), such as postvaccination fatigue, are prevalent following mRNA vaccination, and the underpinnings of which are not understood. Herein, we found that higher baseline expression of genes related to T and NK cell exhaustion and suppression were positively correlated with the development of moderately severe fatigue after Pfizer-BioNTech BNT162b2 vaccination; increased expression of genes associated with T and NK cell exhaustion and suppression reacted to vaccination were associated with greater levels of innate immune activation at 1 day postvaccination. We further found, in a mouse model, that altering the route of vaccination from intramuscular (i.m.) to subcutaneous (s.c.) could lessen the pro-inflammatory response and correspondingly the extent of systemic AEs; the humoral immune response to BNT162b2 vaccination was not compromised. Instead, it is possible that the s.c. route could improve cytotoxic CD8 T-cell responses to BNT162b2 vaccination. Our findings thus provide a glimpse of the molecular basis of postvaccination fatigue from mRNA vaccination and suggest a readily translatable solution to minimize systemic AEs.
Conflict of interest statement
The authors have declared no competing interests exists.
Figures
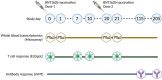

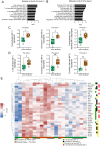
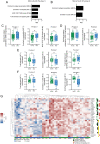

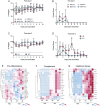

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

