The topoisomerase 3 zinc finger domain cooperates with the RMI1 scaffold to promote stable association of the BTR complex to recombination intermediates in the Caenorhabditis elegans germline
- PMID: 35639927
- PMCID: PMC9178014
- DOI: 10.1093/nar/gkac408
The topoisomerase 3 zinc finger domain cooperates with the RMI1 scaffold to promote stable association of the BTR complex to recombination intermediates in the Caenorhabditis elegans germline
Abstract
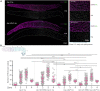
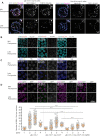
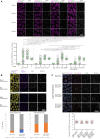
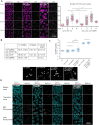
Homologous recombination is the predominant DNA repair pathway used in the gonad. Of the excess DNA double-strand breaks formed in meiosis, only a subset matures into crossovers, with the remainder repaired as non-crossovers. The conserved BTR complex (comprising Bloom helicase, topoisomerase 3 and RMI1/2 scaffold proteins) acts at multiple steps during recombination to dismantle joint DNA molecules, thereby mediating the non-crossover outcome and chromosome integrity. Furthermore, the complex displays a role at the crossover site that is less well understood. Besides catalytic and TOPRIM domains, topoisomerase 3 enzymes contain a variable number of carboxy terminal zinc finger (ZnF) domains. Here, we studied the Caenorhabditis elegans mutant, in which the single ZnF domain is deleted. In contrast to the gene disruption allele, the top-3-ZnF mutant is viable, with no replication defects; the allele appears to be a hypomorph. The TOP-3-ZnF protein is recruited into foci but the mutant has increased numbers of crossovers along its chromosomes, with minor defects in repressing heterologous recombination, and a marked delay in the maturation/processing of recombination intermediates after loading of the RAD-51 recombinase. The ZnF domain cooperates with the RMI1 homolog RMH-2 to stabilize association of the BTR complex with recombination intermediates and to prevent recombination between heterologous DNA sequences.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures