DNMT3A mutations define a unique biological and prognostic subgroup associated with cytotoxic T cells in PTCL-NOS
- PMID: 35639959
- PMCID: PMC9479030
- DOI: 10.1182/blood.2021015019
DNMT3A mutations define a unique biological and prognostic subgroup associated with cytotoxic T cells in PTCL-NOS
Abstract
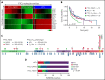
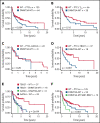
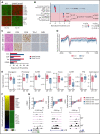
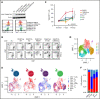
Peripheral T-cell lymphomas (PTCLs) are heterogenous T-cell neoplasms often associated with epigenetic dysregulation. We investigated de novo DNA methyltransferase 3A (DNMT3A) mutations in common PTCL entities, including angioimmunoblastic T-cell lymphoma and novel molecular subtypes identified within PTCL-not otherwise specified (PTCL-NOS) designated as PTCL-GATA3 and PTCL-TBX21. DNMT3A-mutated PTCL-TBX21 cases showed inferior overall survival (OS), with DNMT3A-mutated residues skewed toward the methyltransferase domain and dimerization motif (S881-R887). Transcriptional profiling demonstrated significant enrichment of activated CD8+ T-cell cytotoxic gene signatures in the DNMT3A-mutant PTCL-TBX21 cases, which was further validated using immunohistochemistry. Genomewide methylation analysis of DNMT3A-mutant vs wild-type (WT) PTCL-TBX21 cases demonstrated hypomethylation in target genes regulating interferon-γ (IFN-γ), T-cell receptor signaling, and EOMES (eomesodermin), a master transcriptional regulator of cytotoxic effector cells. Similar findings were observed in a murine model of PTCL with Dnmt3a loss (in vivo) and further validated in vitro by ectopic expression of DNMT3A mutants (DNMT3A-R882, -Q886, and -V716, vs WT) in CD8+ T-cell line, resulting in T-cell activation and EOMES upregulation. Furthermore, stable, ectopic expression of the DNMT3A mutants in primary CD3+ T-cell cultures resulted in the preferential outgrowth of CD8+ T cells with DNMT3AR882H mutation. Single-cell RNA sequencing(RNA-seq) analysis of CD3+ T cells revealed differential CD8+ T-cell subset polarization, mirroring findings in DNMT3A-mutated PTCL-TBX21 and validating the cytotoxic and T-cell memory transcriptional programs associated with the DNMT3AR882H mutation. Our findings indicate that DNMT3A mutations define a cytotoxic subset in PTCL-TBX21 with prognostic significance and thus may further refine pathological heterogeneity in PTCL-NOS and suggest alternative treatment strategies for this subset.
Figures


References
-
- Vose JM. Peripheral T-cell non-Hodgkin’s lymphoma. Hematol Oncol Clin North Am. 2008;22(5):997-1005. - PubMed
-
- Piccaluga PP, Tabanelli V, Pileri SA. Molecular genetics of peripheral T-cell lymphomas. Int J Hematol. 2014;99(3): 219-226. - PubMed
-
- Hsi ED, Horwitz SM, Carson KR, et al. . Analysis of peripheral t-cell lymphoma diagnostic workup in the United States. Clin Lymphoma Myeloma Leuk. 2017;17(4):193-200. - PubMed
-
- Swerdlow SH, Campo E, Harris NL, eds et al.. WHO Classification of tumours of haematopoietic and lymphoid tissues. 4th ed, revised. Lyon, France: International Agency for Research on Cancer; 2017:2
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

