Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins
- PMID: 35641567
- PMCID: PMC9152305
- DOI: 10.1038/s41422-022-00672-4
Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins
Abstract
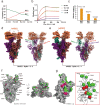
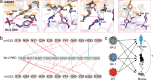
The Omicron BA.2 variant has become a dominant infective strain worldwide. Receptor binding studies show that the Omicron BA.2 spike trimer exhibits 11-fold and 2-fold higher potency in binding to human ACE2 than the spike trimer from the wildtype (WT) and Omicron BA.1 strains. The structure of the BA.2 spike trimer complexed with human ACE2 reveals that all three receptor-binding domains (RBDs) in the spike trimer are in open conformation, ready for ACE2 binding, thus providing a basis for the increased infectivity of the BA.2 strain. JMB2002, a therapeutic antibody that was shown to efficiently inhibit Omicron BA.1, also shows potent neutralization activities against Omicron BA.2. In addition, both BA.1 and BA.2 spike trimers are able to bind to mouse ACE2 with high potency. In contrast, the WT spike trimer binds well to cat ACE2 but not to mouse ACE2. The structures of both BA.1 and BA.2 spike trimer bound to mouse ACE2 reveal the basis for their high affinity interactions. Together, these results suggest a possible evolution pathway for Omicron BA.1 and BA.2 variants via a human-cat-mouse-human circle, which could have important implications in establishing an effective strategy for combating SARS-CoV-2 viral infections.
© 2022. The Author(s).
Conflict of interest statement
H.E.X., W.Y., Y.X., C.W., H.L., M.J., X.X.W., Q.Y., and K.W. have declared no competing interest. S.-J.D., X.C., C.G., J.L., D.W., X.H., and X.P.W., are employees of Shanghai Jemincare Pharmaceuticals Co. Ltd., and are developing JMB2002 as a potential anti-Omicron therapeutic.
Figures





Comment in
-
Structural insights into the Omicron spike trimer: tackling the challenges of continuously evolving SARS-CoV-2 variants.Signal Transduct Target Ther. 2022 Sep 16;7(1):322. doi: 10.1038/s41392-022-01179-5. Signal Transduct Target Ther. 2022. PMID: 36114171 Free PMC article. No abstract available.
Similar articles
-
Characterization of Entry Pathways, Species-Specific Angiotensin-Converting Enzyme 2 Residues Determining Entry, and Antibody Neutralization Evasion of Omicron BA.1, BA.1.1, BA.2, and BA.3 Variants.J Virol. 2022 Sep 14;96(17):e0114022. doi: 10.1128/jvi.01140-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000843 Free PMC article.
-
Evolution of Immune Evasion and Host Range Expansion by the SARS-CoV-2 B.1.1.529 (Omicron) Variant.mBio. 2023 Apr 25;14(2):e0041623. doi: 10.1128/mbio.00416-23. Epub 2023 Apr 3. mBio. 2023. PMID: 37010428 Free PMC article.
-
Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.Science. 2022 Mar 4;375(6584):1048-1053. doi: 10.1126/science.abn8863. Epub 2022 Feb 8. Science. 2022. PMID: 35133176 Free PMC article.
-
Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19.Front Immunol. 2021 Dec 22;12:732690. doi: 10.3389/fimmu.2021.732690. eCollection 2021. Front Immunol. 2021. PMID: 35003058 Free PMC article. Review.
-
The emergence of SARS-CoV-2 Omicron subvariants: current situation and future trends.Infez Med. 2022 Dec 1;30(4):480-494. doi: 10.53854/liim-3004-2. eCollection 2022. Infez Med. 2022. PMID: 36482957 Free PMC article. Review.
Cited by
-
SARS-CoV-2 exposure in Norway rats (Rattus norvegicus) from New York City.bioRxiv [Preprint]. 2022 Dec 10:2022.11.18.517156. doi: 10.1101/2022.11.18.517156. bioRxiv. 2022. Update in: mBio. 2023 Apr 25;14(2):e0362122. doi: 10.1128/mbio.03621-22. PMID: 36451891 Free PMC article. Updated. Preprint.
-
SARS-CoV-2 Exposure in Norway Rats (Rattus norvegicus) from New York City.mBio. 2023 Apr 25;14(2):e0362122. doi: 10.1128/mbio.03621-22. Epub 2023 Mar 9. mBio. 2023. PMID: 36892291 Free PMC article.
-
Exploring Conformational Landscapes and Cryptic Binding Pockets in Distinct Functional States of the SARS-CoV-2 Omicron BA.1 and BA.2 Trimers: Mutation-Induced Modulation of Protein Dynamics and Network-Guided Prediction of Variant-Specific Allosteric Binding Sites.Viruses. 2023 Sep 27;15(10):2009. doi: 10.3390/v15102009. Viruses. 2023. PMID: 37896786 Free PMC article.
-
Probing conformational landscapes of binding and allostery in the SARS-CoV-2 omicron variant complexes using microsecond atomistic simulations and perturbation-based profiling approaches: hidden role of omicron mutations as modulators of allosteric signaling and epistatic relationships.Phys Chem Chem Phys. 2023 Aug 16;25(32):21245-21266. doi: 10.1039/d3cp02042h. Phys Chem Chem Phys. 2023. PMID: 37548589 Free PMC article.
-
Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein.Nat Commun. 2024 Feb 29;15(1):1854. doi: 10.1038/s41467-024-46104-2. Nat Commun. 2024. PMID: 38424106 Free PMC article.
References
-
- Genomic epidemiology of SARS-CoV-2 with global subsampling, https://nextstrain.org/ncov/ (2022).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

