Statistical power for cluster analysis
- PMID: 35641905
- PMCID: PMC9158113
- DOI: 10.1186/s12859-022-04675-1
Statistical power for cluster analysis
Abstract
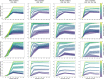
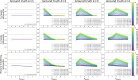
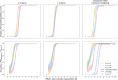
Background: Cluster algorithms are gaining in popularity in biomedical research due to their compelling ability to identify discrete subgroups in data, and their increasing accessibility in mainstream software. While guidelines exist for algorithm selection and outcome evaluation, there are no firmly established ways of computing a priori statistical power for cluster analysis. Here, we estimated power and classification accuracy for common analysis pipelines through simulation. We systematically varied subgroup size, number, separation (effect size), and covariance structure. We then subjected generated datasets to dimensionality reduction approaches (none, multi-dimensional scaling, or uniform manifold approximation and projection) and cluster algorithms (k-means, agglomerative hierarchical clustering with Ward or average linkage and Euclidean or cosine distance, HDBSCAN). Finally, we directly compared the statistical power of discrete (k-means), "fuzzy" (c-means), and finite mixture modelling approaches (which include latent class analysis and latent profile analysis).
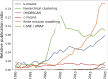
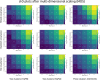
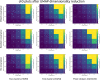
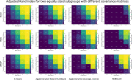
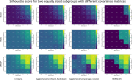
Results: We found that clustering outcomes were driven by large effect sizes or the accumulation of many smaller effects across features, and were mostly unaffected by differences in covariance structure. Sufficient statistical power was achieved with relatively small samples (N = 20 per subgroup), provided cluster separation is large (Δ = 4). Finally, we demonstrated that fuzzy clustering can provide a more parsimonious and powerful alternative for identifying separable multivariate normal distributions, particularly those with slightly lower centroid separation (Δ = 3).
Conclusions: Traditional intuitions about statistical power only partially apply to cluster analysis: increasing the number of participants above a sufficient sample size did not improve power, but effect size was crucial. Notably, for the popular dimensionality reduction and clustering algorithms tested here, power was only satisfactory for relatively large effect sizes (clear separation between subgroups). Fuzzy clustering provided higher power in multivariate normal distributions. Overall, we recommend that researchers (1) only apply cluster analysis when large subgroup separation is expected, (2) aim for sample sizes of N = 20 to N = 30 per expected subgroup, (3) use multi-dimensional scaling to improve cluster separation, and (4) use fuzzy clustering or mixture modelling approaches that are more powerful and more parsimonious with partially overlapping multivariate normal distributions.
Keywords: Cluster analysis; Covariance; Dimensionality reduction; Effect size; Latent class analysis; Latent profile analysis; Sample size; Simulation; Statistical power.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830.
-
- Hennig C. fpc. 2020. Available from https://cran.r-project.org/web/packages/fpc/index.html.
-
- Anjana RM, Baskar V, Nair ATN, Jebarani S, Siddiqui MK, Pradeepa R, et al. Novel subgroups of type 2 diabetes and their association with microvascular outcomes in an Asian Indian population: a data-driven cluster analysis: the INSPIRED study. BMJ Open Diabetes Res Care. 2020;8(1):e001506. doi: 10.1136/bmjdrc-2020-001506. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

