Genetic Variation in Reproductive Investment Across an Ephemerality Gradient in Daphnia pulex
- PMID: 35642301
- PMCID: PMC9198359
- DOI: 10.1093/molbev/msac121
Genetic Variation in Reproductive Investment Across an Ephemerality Gradient in Daphnia pulex
Abstract
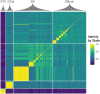
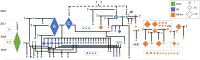
Species across the tree of life can switch between asexual and sexual reproduction. In facultatively sexual species, the ability to switch between reproductive modes is often environmentally dependent and subject to local adaptation. However, the ecological and evolutionary factors that influence the maintenance and turnover of polymorphism associated with facultative sex remain unclear. We studied the ecological and evolutionary dynamics of reproductive investment in the facultatively sexual model species, Daphnia pulex. We found that patterns of clonal diversity, but not genetic diversity varied among ponds consistent with the predicted relationship between ephemerality and clonal structure. Reconstruction of a multi-year pedigree demonstrated the coexistence of clones that differ in their investment into male production. Mapping of quantitative variation in male production using lab-generated and field-collected individuals identified multiple putative quantitative trait loci (QTL) underlying this trait, and we identified a plausible candidate gene. The evolutionary history of these QTL suggests that they are relatively young, and male limitation in this system is a rapidly evolving trait. Our work highlights the dynamic nature of the genetic structure and composition of facultative sex across space and time and suggests that quantitative genetic variation in reproductive strategy can undergo rapid evolutionary turnover.
Keywords: Daphnia; facultative sex; male production; population genomics; quantitative genetics.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures




References
-
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw. 67:1–48.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

