Natural allelic variation of GmST05 controlling seed size and quality in soybean
- PMID: 35642379
- PMCID: PMC9398382
- DOI: 10.1111/pbi.13865
Natural allelic variation of GmST05 controlling seed size and quality in soybean
Abstract
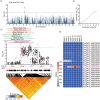
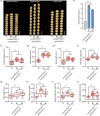
Seed size is one of the most important agronomic traits determining the yield of crops. Cloning the key genes controlling seed size and pyramiding their elite alleles will facilitate yield improvement. To date, few genes controlling seed size have been identified in soybean, a major crop that provides half of the plant oil and one quarter of the plant protein globally. Here, through a genome-wide association study of over 1800 soybean accessions, we determined that natural allelic variation at GmST05 (Seed Thickness 05) predominantly controlled seed thickness and size in soybean germplasm. Further analyses suggested that the two major haplotypes of GmST05 differed significantly at the transcriptional level. Transgenic experiments demonstrated that GmST05 positively regulated seed size and influenced oil and protein contents, possibly by regulating the transcription of GmSWEET10a. Population genetic diversity analysis suggested that allelic variations of GmST05 were selected during geographical differentiation but have not been fixed. In summary, natural variation in GmST05 determines transcription levels and influences seed size and quality in soybean, making it an important gene resource for soybean molecular breeding.
Keywords: GmST05; GWAS; natural allelic variation; oil and protein contents; seed size; soybean.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bai, X. , Huang, Y. , Hu, Y. , Liu, H. , Zhang, B. , Smaczniak, C. , Hu, G. et al. (2017) Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants, 3, 885–893. - PubMed
-
- Bednarek, J. , Boulaflous, A. , Girousse, C. , Ravel, C. , Tassy, C. , Barret, P. , Bouzidi, M.F. et al. (2012) Down‐regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 63, 5945–5955. - PubMed
-
- Bradley, D. , Ratcliffe, O. , Vincent, C. , Carpenter, R. and Coen, E. (1997) Inflorescence commitment and architecture in Arabidopsis . Science, 275, 80–83. - PubMed
-
- Brummer, E. , Graef, G. , Orf, J. , Wilcox, J. and Shoemaker, R. (1997) Mapping QTL for seed protein and oil content in eight soybean populations. Crop. Sci. 37, 370–378.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

