Analyzing and Modeling the Spread of SARS-CoV-2 Omicron Lineages BA.1 and BA.2, France, September 2021-February 2022
- PMID: 35642476
- PMCID: PMC9239895
- DOI: 10.3201/eid2807.220033
Analyzing and Modeling the Spread of SARS-CoV-2 Omicron Lineages BA.1 and BA.2, France, September 2021-February 2022
Abstract
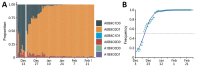
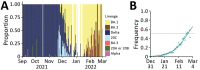
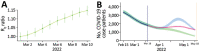
We analyzed 324,734 SARS-CoV-2 variant screening tests from France enriched with 16,973 whole-genome sequences sampled during September 1, 2021-February 28, 2022. Results showed the estimated growth advantage of the Omicron variant over the Delta variant to be 105% (95% CI 96%-114%) and that of the BA.2 lineage over the BA.1 lineage to be 49% (95% CI 44%-52%). Quantitative PCR cycle threshold values were consistent with an increased ability of Omicron to generate breakthrough infections. Epidemiologic modeling shows that, in spite of its decreased virulence, the Omicron variant can generate important critical COVID-19 activity in hospitals in France. The magnitude of the BA.2 wave in hospitals depends on the level of relaxing of control measures but remains lower than that of BA.1 in median scenarios.
Keywords: BA.1; BA.2; COVID-19; France; Omicron; PCR; SARS-CoV-2; coronavirus disease; epidemiology; genomics; mass screening; mutation; respiratory infections; severe acute respiratory syndrome coronavirus 2; vaccine-preventable diseases; variants; viruses; zoonoses.
Figures
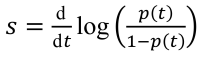
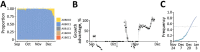


References
-
- WHO Collaborating Centre for Infectious Disease Modelling, MRC Centre for Global Infectious Disease Analysis, Jameel Institute, Imperial College London. Report 50: hospitalisation risk for Omicron cases in England. 2021. Dec 21 [cited 2022 May 6]. https://www.imperial.ac.uk/medicine/departments/school-public-health/inf...
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

