Predicting time-resolved electrophysiological brain networks from structural eigenmodes
- PMID: 35642600
- PMCID: PMC9435022
- DOI: 10.1002/hbm.25967
Predicting time-resolved electrophysiological brain networks from structural eigenmodes
Abstract
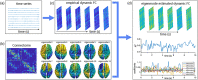
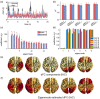
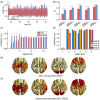
How temporal modulations in functional interactions are shaped by the underlying anatomical connections remains an open question. Here, we analyse the role of structural eigenmodes, in the formation and dissolution of temporally evolving functional brain networks using resting-state magnetoencephalography and diffusion magnetic resonance imaging data at the individual subject level. Our results show that even at short timescales, phase and amplitude connectivity can partly be expressed by structural eigenmodes, but hardly by direct structural connections. Albeit a stronger relationship was found between structural eigenmodes and time-resolved amplitude connectivity. Time-resolved connectivity for both phase and amplitude was mostly characterised by a stationary process, superimposed with very brief periods that showed deviations from this stationary process. For these brief periods, dynamic network states were extracted that showed different expressions of eigenmodes. Furthermore, the eigenmode expression was related to overall cognitive performance and co-occurred with fluctuations in community structure of functional networks. These results implicate that ongoing time-resolved resting-state networks, even at short timescales, can to some extent be understood in terms of activation and deactivation of structural eigenmodes and that these eigenmodes play a role in the dynamic integration and segregation of information across the cortex, subserving cognitive functions.
Keywords: dynamic functional connectivity; eigenmodes; magnetoencephalography.
© 2022 The Authors. Human Brain Mapping published by Wiley Periodicals LLC.
Conflict of interest statement
There were no competing or conflicting interests for any of the authors.
Figures


References
-
- Andersson, C. A. , & Bro, R. (2000). The N‐way toolbox for MATLAB. Chemometrics and Intelligent Laboratory Systems, 52, 1–4.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

