SIK2 inhibition enhances PARP inhibitor activity synergistically in ovarian and triple-negative breast cancers
- PMID: 35642638
- PMCID: PMC9151707
- DOI: 10.1172/JCI146471
SIK2 inhibition enhances PARP inhibitor activity synergistically in ovarian and triple-negative breast cancers
Abstract
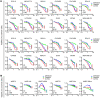
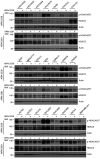
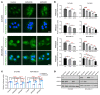
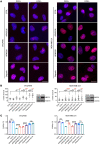
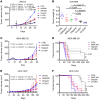
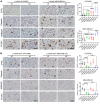
Poly(ADP-ribose) polymerase inhibitors (PARP inhibitors) have had an increasing role in the treatment of ovarian and breast cancers. PARP inhibitors are selectively active in cells with homologous recombination DNA repair deficiency caused by mutations in BRCA1/2 and other DNA repair pathway genes. Cancers with homologous recombination DNA repair proficiency respond poorly to PARP inhibitors. Cancers that initially respond to PARP inhibitors eventually develop drug resistance. We have identified salt-inducible kinase 2 (SIK2) inhibitors, ARN3236 and ARN3261, which decreased DNA double-strand break (DSB) repair functions and produced synthetic lethality with multiple PARP inhibitors in both homologous recombination DNA repair deficiency and proficiency cancer cells. SIK2 is required for centrosome splitting and PI3K activation and regulates cancer cell proliferation, metastasis, and sensitivity to chemotherapy. Here, we showed that SIK2 inhibitors sensitized ovarian and triple-negative breast cancer (TNBC) cells and xenografts to PARP inhibitors. SIK2 inhibitors decreased PARP enzyme activity and phosphorylation of class-IIa histone deacetylases (HDAC4/5/7). Furthermore, SIK2 inhibitors abolished class-IIa HDAC4/5/7-associated transcriptional activity of myocyte enhancer factor-2D (MEF2D), decreasing MEF2D binding to regulatory regions with high chromatin accessibility in FANCD2, EXO1, and XRCC4 genes, resulting in repression of their functions in the DNA DSB repair pathway. The combination of PARP inhibitors and SIK2 inhibitors provides a therapeutic strategy to enhance PARP inhibitor sensitivity for ovarian cancer and TNBC.
Keywords: Apoptosis; Cancer; Cell Biology; DNA repair; Therapeutics.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

