How inclusive are cell lines in preclinical engineered cancer models?
- PMID: 35642685
- PMCID: PMC9187871
- DOI: 10.1242/dmm.049520
How inclusive are cell lines in preclinical engineered cancer models?
Abstract
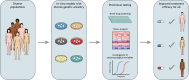
Diverse factors contribute to significant and dire disparities in cancer risk and treatment outcomes. To address this, there was a call for inclusion of sex as a biological variable, which resulted in more instances of careful inclusion of sex in preclinical studies of cancer. Another variable in cancer treatment is genetic ancestry. Although this is considered explicitly in clinical research, it is considerably neglected in preclinical studies. Preclinical research can use several 3D in vitro model systems, such as spheroids/organoids, xenografts, or other bioengineered systems that combine biomaterials and cellular material. Ultimately, the cellular base for all of these in vitro model systems is derived from human cell lines or patient samples, to investigate mechanisms of cancer and screen novel therapeutics, all of which aim to maximize successful outcomes in clinical trials. This in itself offers an opportunity to potentiate effective treatments for many groups of people, when diverse variables like genetic ancestry are consciously included into study design. This Perspective highlights the need for conscious inclusion of genetic ancestry in preclinical cancer tissue engineering, especially when it pertains to determining therapeutic outcomes.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The author declares no competing or financial interests.
Figures
References
-
- Abimiku, A. G., Croxton, T., Ozumba, P. J., Agala, N., Balogun, O., Jonathan, E., Onyemata, E., Ndifon, K., Nadoma, S., Anazodo, T.et al. (2019). Blueprint for building a biorepository in a resource-limited setting that follows international best practices. Afr. J. Lab. Med. 8, a722. 10.4102/ajlm.v8i1.722 - DOI - PMC - PubMed
-
- Agarwal, S., Chakravarthi, B. V. S. K., Behring, M., Kim, H. G., Chandrashekar, D. S., Gupta, N., Bajpai, P., Elkholy, A., Balasubramanya, S. A. H., Hardy, C.et al. (2020). PAICS, a purine nucleotide metabolic enzyme, is involved in tumor growth and the metastasis of colorectal cancer. Cancers 12, 772. 10.3390/cancers12040772 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

