Clonal evolution after treatment pressure in multiple myeloma: heterogenous genomic aberrations and transcriptomic convergence
- PMID: 35643867
- PMCID: PMC9252918
- DOI: 10.1038/s41375-022-01597-y
Clonal evolution after treatment pressure in multiple myeloma: heterogenous genomic aberrations and transcriptomic convergence
Abstract
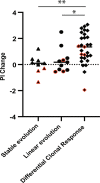
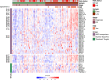
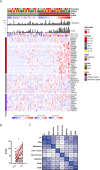
We investigated genomic and transcriptomic changes in paired tumor samples of 29 in-house multiple myeloma (MM) patients and 28 patients from the MMRF CoMMpass study before and after treatment. A change in clonal composition was found in 46/57 (82%) of patients, and single-nucleotide variants (SNVs) increased from median 67 to 86. The highest increase in prevalence of genetic aberrations was found in RAS genes (60% to 72%), amp1q21 (18% to 35%), and TP53 (9% to 18%). The SBS-MM1 mutation signature was detected both in patients receiving high and low dose melphalan. A total of 2589 genes were differentially expressed between early and late samples (FDR < 0.05). Gene set enrichment analysis (GSEA) showed increased expression of E2F, MYC, and glycolysis pathways and a decreased expression in TNF-NFkB and TGFbeta pathways in late compared to early stage. Single sample GSEA (ssGSEA) scores of differentially expressed pathways revealed that these changes were most evident in end-stage disease. Increased expression of several potentially targetable genes was found at late disease stages, including cancer-testis antigens, XPO1 and ABC transporters. Our study demonstrates a transcriptomic convergence of pathways supporting increased proliferation and metabolism during disease progression in MM.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

