Noninvasive prenatal prediction of fetal haplotype with Spearman rank correlation analysis model
- PMID: 35644943
- PMCID: PMC9356545
- DOI: 10.1002/mgg3.1988
Noninvasive prenatal prediction of fetal haplotype with Spearman rank correlation analysis model
Abstract
Background: Noninvasive prenatal testing (NIPT) has been widely used clinically to detect fetal chromosomal aneuploidy with high accuracy rates, gradually replacing traditional serological screening. However, the application of NIPT for monogenic diseases is still in an immature stage of exploration. The detection of mutations in peripheral blood of pregnant women requires precise qualitative and quantitative techniques, which limits its application. The bioinformatic strategies based on the SNP (single nucleotide polymorphism) linkage analysis are more practical, which can be divided into two types depending on whether proband information is needed. Hidden Markov Mode (HMM) and Sequential probability ratio test (SPRT) are suitable for families with probands. In contrast, methods based on databases and population demographic information are suitable for families without probands.
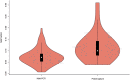
Methods: In this study, we proposed a Spearman rank correlation analysis method to infer the fetal haplotypes based on core family information. Allele frequencies of SNPs that were used to construct parental haplotypes were calculated as sets of nonparametric variables, in contrast to their theoretical values represented by a fetal fraction (FF). The effects on the calculation of the fetal concentration of two DNA enrichment methods, multiple-PCR amplification, and targeted hybrid capture, were compared, and the heterozygosity distribution of SNPs within pedigrees was analyzed to reveal the best conditions for the model application.
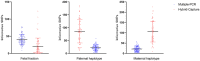
Results: Predictions of the paternal haplotype inheritance were in line with expectations for both DNA library construction methods, while for maternal haplotype inheritance prediction, the rates were 96.55% for method multiple-PCR amplification and 95.8% for method targeted hybrid capture.
Conclusion: Positive prediction rates showed that the maternal haplotype prediction was not as accurate as paternal one, due to the large amount of maternal noise in the mother's peripheral blood. Although this model is relatively immature, it provides a new perspective for noninvasive prenatal clinical tests of monogenic diseases.
Keywords: Spearman rank correlation analysis model; haplotype; multiple-PCR; targeted hybrid capture.
© 2022 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that they have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service, and/or company that could be construed as influencing the manuscript entitled.
Figures


References
-
- Ameur, A. , Kloosterman, W. P. , & Hestand, M. S. (2019). Single‐molecule sequencing: Towards clinical applications. Trends in Biotechnology, 37(1), 72–85. - PubMed
-
- Barrett, A. N. , McDonnell, T. C. R. , Chan, K. C. A. , & Chitty, L. S. (2012). Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clinical Chemistry, 58(6), 1026–1032. - PubMed
-
- Chan, K. A. , Jiang, P. , Sun, K. , Cheng, Y. K. , Tong, Y. K. , Cheng, S. H. , Wong, A. I. , Hudecova, I. , Leung, T. Y. , Chiu, R. W. , & Lo, Y. M. (2016). Second generation noninvasive fetal genome analysis reveals de novo mutations, single‐base parental inheritance, and preferred DNA ends. Proceedings of the National Academy of Sciences of the United States of America, 113(50), E8159–E8168. - PMC - PubMed
-
- Chen, C. , Li, R. , Sun, J. , Zhu, Y. , Jiang, L. , Li, J. , Fu, F. , Wan, J. , Guo, F. , An, X. , Wang, Y. , Fan, L. , Sun, Y. , Guo, X. , Zhao, S. , Wang, W. , Zeng, F. , Yang, Y. , Ni, P. , … Liao, C. (2021). Noninvasive prenatal testing of alpha‐thalassemia and beta‐thalassemia through population‐based parental haplotyping. Genome Medicine, 13(1), 18. - PMC - PubMed
-
- Duan, H. , Liu, N. , Zhao, Z. , Liu, Y. , Wang, Y. , Li, Z. , Xu, M. , Cram, D. S. , & Kong, X. (2019). Non‐invasive prenatal testing of pregnancies at risk for phenylketonuria. Archives of Disease in Childhood. Fetal and Neonatal Edition, 104(1), F24–F29. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

