Heterologous Expression of Arabidopsis AtARA6 in Soybean Enhances Salt Tolerance
- PMID: 35646070
- PMCID: PMC9134241
- DOI: 10.3389/fgene.2022.849357
Heterologous Expression of Arabidopsis AtARA6 in Soybean Enhances Salt Tolerance
Erratum in
-
Erratum: Heterologous expression of Arabidopsis AtARA6 in soybean enhances salt tolerance.Front Genet. 2022 Sep 8;13:1017834. doi: 10.3389/fgene.2022.1017834. eCollection 2022. Front Genet. 2022. PMID: 36159978 Free PMC article.
Abstract
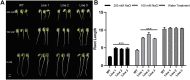
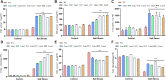
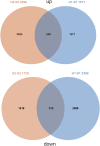
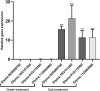
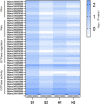
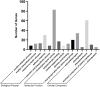
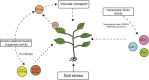
Salt damage is an important abiotic stress affecting the agronomic traits of soybean. Soybeans rapidly sense and transmit adverse signals when salt-damaged, inducing a set of response mechanisms to resist salt stress. AtARA6 encodes a small GTPase, which plays an important role in Arabidopsis vesicle transport and salt tolerance. In this study, we transformed the Arabidopsis gene AtARA6 into the cultivated soybean Shen Nong 9 (SN9). To investigate the salt tolerance pathways affected by AtARA6 in soybean, we performed transcriptome sequencing using transgenic soybean and wild-type (SN9) under salt treatment and water treatment. Our results suggest that AtARA6 is involved in the regulation of soybean SNARE complexes in the vesicle transport pathway, which may directly strengthen salt tolerance. In addition, we comprehensively analyzed the RNA-seq data of transgenic soybean and SN9 under different treatments and obtained 935 DEGs. GO analysis showed that these DEGs were significantly enriched in transcription factor activity, sequence-specific DNA binding, and the inositol catabolic process. Three salt-responsive negative regulator transcription factors, namely MYC2, WRKY6, and WRKY86, were found to be significantly downregulated after salt treatment in transgenic soybeans. Moreover, four genes encoding inositol oxygenase were significantly enriched in the inositol catabolic process pathway, which could improve the salt tolerance of transgenic soybeans by reducing their reactive oxygen species content. These are unique salt tolerance effects produced by transgenic soybeans. Our results provide basic insights into the function of AtARA6 in soybeans and its role in abiotic stress processes in plants.
Keywords: AtARA6; RAB GTPase; RAB5; SNARE pathway; salt tolerance; sybean.
Copyright © 2022 Hong, Li, Zhao, Yang, Zhang, Teng, Jing, Kong, Liu, Li, Meng, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Erratum: Heterologous expression of Arabidopsis AtARA6 in soybean enhances salt tolerance.Front Genet. 2022 Sep 8;13:1017834. doi: 10.3389/fgene.2022.1017834. eCollection 2022. Front Genet. 2022. PMID: 36159978 Free PMC article.
-
Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana.J Exp Bot. 2016 Jan;67(1):175-94. doi: 10.1093/jxb/erv450. Epub 2015 Oct 14. J Exp Bot. 2016. PMID: 26466661
-
A Glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher Na+ efflux rate and K+/Na+ ratio in Arabidopsis.BMC Plant Biol. 2019 Nov 5;19(1):469. doi: 10.1186/s12870-019-2084-4. BMC Plant Biol. 2019. PMID: 31690290 Free PMC article.
-
Overexpression of an aquaporin protein from Aspergillus glaucus confers salt tolerance in transgenic soybean.Transgenic Res. 2021 Dec;30(6):727-737. doi: 10.1007/s11248-021-00280-9. Epub 2021 Aug 30. Transgenic Res. 2021. PMID: 34460070 Review.
-
Salt tolerance in soybean.J Integr Plant Biol. 2008 Oct;50(10):1196-212. doi: 10.1111/j.1744-7909.2008.00760.x. J Integr Plant Biol. 2008. PMID: 19017107 Review.
Cited by
-
Overexpression of the aldehyde dehydrogenase AhALDH3H1 from Arachis hypogaea in soybean increases saline-alkali stress tolerance.Front Plant Sci. 2023 Mar 28;14:1165384. doi: 10.3389/fpls.2023.1165384. eCollection 2023. Front Plant Sci. 2023. PMID: 37056489 Free PMC article.
-
Unfolding molecular switches for salt stress resilience in soybean: recent advances and prospects for salt-tolerant smart plant production.Front Plant Sci. 2023 Apr 19;14:1162014. doi: 10.3389/fpls.2023.1162014. eCollection 2023. Front Plant Sci. 2023. PMID: 37152141 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

