Multi-Size Deep Learning Based Preoperative Computed Tomography Signature for Prognosis Prediction of Colorectal Cancer
- PMID: 35646105
- PMCID: PMC9133721
- DOI: 10.3389/fgene.2022.880093
Multi-Size Deep Learning Based Preoperative Computed Tomography Signature for Prognosis Prediction of Colorectal Cancer
Abstract
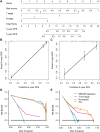
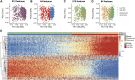
Background: Preoperative and postoperative evaluation of colorectal cancer (CRC) patients is crucial for subsequent treatment guidance. Our study aims to provide a timely and rapid assessment of the prognosis of CRC patients with deep learning according to non-invasive preoperative computed tomography (CT) and explore the underlying biological explanations. Methods: A total of 808 CRC patients with preoperative CT (development cohort: n = 426, validation cohort: n = 382) were enrolled in our study. We proposed a novel end-to-end Multi-Size Convolutional Neural Network (MSCNN) to predict the risk of CRC recurrence with CT images (CT signature). The prognostic performance of CT signature was evaluated by Kaplan-Meier curve. An integrated nomogram was constructed to improve the clinical utility of CT signature by combining with other clinicopathologic factors. Further visualization and correlation analysis for CT deep features with paired gene expression profiles were performed to reveal the molecular characteristics of CRC tumors learned by MSCNN in radiographic imaging. Results: The Kaplan-Meier analysis showed that CT signature was a significant prognostic factor for CRC disease-free survival (DFS) prediction [development cohort: hazard ratio (HR): 50.7, 95% CI: 28.4-90.6, p < 0.001; validation cohort: HR: 2.04, 95% CI: 1.44-2.89, p < 0.001]. Multivariable analysis confirmed the independence prognostic value of CT signature (development cohort: HR: 30.7, 95% CI: 19.8-69.3, p < 0.001; validation cohort: HR: 1.83, 95% CI: 1.19-2.83, p = 0.006). Dimension reduction and visualization of CT deep features demonstrated a high correlation with the prognosis of CRC patients. Functional pathway analysis further indicated that CRC patients with high CT signature presented down-regulation of several immunology pathways. Correlation analysis found that CT deep features were mainly associated with activation of metabolic and proliferative pathways. Conclusions: Our deep learning based preoperative CT signature can effectively predict prognosis of CRC patients. Integration analysis of multi-omic data revealed that some molecular characteristics of CRC tumor can be captured by deep learning in CT images.
Keywords: colorectal cancer; deep learning; nomogram; pathway analysis; prognosis.
Copyright © 2022 Li, Cai, Zhong, Lv, Huang, Zhu, Hu, Qi, Wu and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A Metabolism-Related Radiomics Signature for Predicting the Prognosis of Colorectal Cancer.Front Mol Biosci. 2021 Jan 7;7:613918. doi: 10.3389/fmolb.2020.613918. eCollection 2020. Front Mol Biosci. 2021. PMID: 33490106 Free PMC article.
-
Development and Validation of a Deep Learning CT Signature to Predict Survival and Chemotherapy Benefit in Gastric Cancer: A Multicenter, Retrospective Study.Ann Surg. 2021 Dec 1;274(6):e1153-e1161. doi: 10.1097/SLA.0000000000003778. Ann Surg. 2021. PMID: 31913871
-
A Nomogram Based on CT Deep Learning Signature: A Potential Tool for the Prediction of Overall Survival in Resected Non-Small Cell Lung Cancer Patients.Cancer Manag Res. 2021 Mar 30;13:2897-2906. doi: 10.2147/CMAR.S299020. eCollection 2021. Cancer Manag Res. 2021. PMID: 33833572 Free PMC article.
-
Non-invasive prediction of microsatellite instability in colorectal cancer by a genetic algorithm-enhanced artificial neural network-based CT radiomics signature.Eur Radiol. 2023 Jan;33(1):11-22. doi: 10.1007/s00330-022-08954-6. Epub 2022 Jun 30. Eur Radiol. 2023. PMID: 35771245
-
Cancer-associated fibroblasts impact the clinical outcome and treatment response in colorectal cancer via immune system modulation: a comprehensive genome-wide analysis.Mol Med. 2021 Oct 30;27(1):139. doi: 10.1186/s10020-021-00402-3. Mol Med. 2021. PMID: 34717544 Free PMC article.
Cited by
-
Artificial Intelligence Applications in the Treatment of Colorectal Cancer: A Narrative Review.Clin Med Insights Oncol. 2024 Jan 5;18:11795549231220320. doi: 10.1177/11795549231220320. eCollection 2024. Clin Med Insights Oncol. 2024. PMID: 38187459 Free PMC article. Review.
-
Research status and trends of deep learning in colorectal cancer (2011-2023): Bibliometric analysis and visualization.World J Gastrointest Oncol. 2025 May 15;17(5):103667. doi: 10.4251/wjgo.v17.i5.103667. World J Gastrointest Oncol. 2025. PMID: 40487952 Free PMC article.
-
Fully automated 3D multi-modal deep learning model for preoperative T-stage prediction of colorectal cancer using 18F-FDG PET/CT.Eur J Nucl Med Mol Imaging. 2025 Jul 28. doi: 10.1007/s00259-025-07450-5. Online ahead of print. Eur J Nucl Med Mol Imaging. 2025. PMID: 40719866
-
Population-based colorectal cancer risk prediction using a SHAP-enhanced LightGBM model.Front Oncol. 2025 Jul 17;15:1575844. doi: 10.3389/fonc.2025.1575844. eCollection 2025. Front Oncol. 2025. PMID: 40746597 Free PMC article.
-
Deep Learning of radiology-genomics integration for computational oncology: A mini review.Comput Struct Biotechnol J. 2024 Jun 20;23:2708-2716. doi: 10.1016/j.csbj.2024.06.019. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39035833 Free PMC article. Review.
References
-
- Bilal M., Raza S. E. A., Azam A., Graham S., Ilyas M., Cree I. A., et al. (2021). Development and Validation of a Weakly Supervised Deep Learning Framework to Predict the Status of Molecular Pathways and Key Mutations in Colorectal Cancer from Routine Histology Images: a Retrospective Study. Lancet Digital Health 3 (12), e763–e772. 10.1016/s2589-7500(21)00180-1 - DOI - PMC - PubMed
-
- He K., Zhang X., Ren S., Sun J. (2016). “Deep Residual Learning for Image Recognition,” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27-30 June 2016, 770–778.
LinkOut - more resources
Full Text Sources

