Dynamic Transcriptome Profiling Reveals LncRNA-Centred Regulatory Networks in the Modulation of Pluripotency
- PMID: 35646895
- PMCID: PMC9130768
- DOI: 10.3389/fcell.2022.880674
Dynamic Transcriptome Profiling Reveals LncRNA-Centred Regulatory Networks in the Modulation of Pluripotency
Abstract
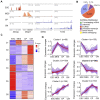
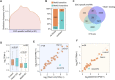
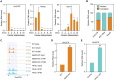
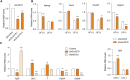
Long noncoding RNAs (lncRNAs) have emerged as vital regulators of gene expression during embryonic stem cell (ESC) self-renewal and differentiation. Here, we systemically analyzed the differentially regulated lncRNAs during ESC-derived cardiomyocyte (CM) differentiation. We established a perspicuous profile of lncRNA expression at four critical developmental stages and found that the differentially expressed lncRNAs were grouped into six distinct clusters. The cluster with specific expression in ESC enriches the largest number of lncRNAs. Investigation of lncRNA-protein interaction network revealed that they are not only controlled by classic key transcription factors, but also modulated by epigenetic and epitranscriptomic factors including N6-methyladenosine (m6A) effector machineries. A detailed inspection revealed that 28 out of 385 lncRNAs were modified by methylation as well as directly recruited by the nuclear m6A reader protein Ythdc1. Unlike other 27 non-coding transcripts, the ESC-specific lncRNA Gm2379, located in both nucleus and cytoplasm, becomes dramatically upregulated in response to the depletion of m6A or Ythdc1. Consistent with the role of m6A in cell fate regulation, depletion of Gm2379 results in dysregulated expressions of pluripotent genes and crucial genes required for the formation of three germ layers. Collectively, our study provides a foundation for understanding the dynamic regulation of lncRNA transcriptomes during ESC differentiation and identifies the interplay between epitranscriptomic modification and key lncRNAs in the regulation of cell fate decision.
Keywords: Gm2379; cell fate; embryonic stem cell; lncRNA; m6A modificaiton.
Copyright © 2022 Wang, Zhang, Ding, Zhang, Wu, Huang, He, Zhou and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

