Nuclear GSK-3β and Oncogenic KRas Lead to the Retention of Pancreatic Ductal Progenitor Cells Phenotypically Similar to Those Seen in IPMN
- PMID: 35646902
- PMCID: PMC9136019
- DOI: 10.3389/fcell.2022.853003
Nuclear GSK-3β and Oncogenic KRas Lead to the Retention of Pancreatic Ductal Progenitor Cells Phenotypically Similar to Those Seen in IPMN
Abstract
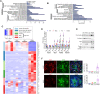
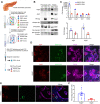
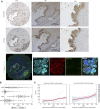
Glycogen synthase kinase-3β (GSK-3β) is a downstream target of oncogenic KRas and can accumulate in the nucleus in pancreatic ductal adenocarcinoma (PDA). To determine the interplay between oncogenic KRas and nuclear GSK-3β in PDA development, we generated Lox-STOP-Lox (LSL) nuclear-targeted GSK-3β animals and crossed them with LSL-KRasG12D mice under the control of the Pdx1-cre transgene-referred to as KNGC. Interestingly, 4-week-old KNGC animals show a profound loss of acinar cells, the expansion of ductal cells, and the rapid development of cystic-like lesions reminiscent of intraductal papillary mucinous neoplasm (IPMN). RNA-sequencing identified the expression of several ductal cell lineage genes including AQP5. Significantly, the Aqp5+ ductal cell pool was proliferative, phenotypically distinct from quiescent pancreatic ductal cells, and deletion of AQP5 limited expansion of the ductal pool. Aqp5 is also highly expressed in human IPMN along with GSK-3β highlighting the putative role of Aqp5+ ductal cells in human preneoplastic lesion development. Altogether, these data identify nGSK-3β and KRasG12D as an important signaling node promoting the retention of pancreatic ductal progenitor cells, which could be used to further characterize pancreatic ductal development as well as lineage biomarkers related to IPMN and PDA.
Keywords: AQP5; GSK-3β; KRAS; intraductal papillary mucinous neoplasm; pancreatic adenocarcinoma; progenitor cell.
Copyright © 2022 Ding, Roeck, Zhang, Zidek, Rodman, Hernandez-Barco, Zhang, Bamlet, Oberg, Zhang, Bardeesy, Li and Billadeau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- American Cancer Society (2019). Cancer Facts & Figures 2019. Atlanta: American Cancer Society.
-
- Auciello F. R., Bulusu V., Oon C., Tait-Mulder J., Berry M., Bhattacharyya S., et al. (2019). A Stromal Lysolipid-Autotaxin Signaling Axis Promotes Pancreatic Tumor Progression. Cancer Discov. 9, 617–627. 10.1158/2159-8290.cd-18-1212 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

