Synthetic Heparan Sulfate Mimetic Pixatimod (PG545) Potently Inhibits SARS-CoV-2 by Disrupting the Spike-ACE2 Interaction
- PMID: 35647275
- PMCID: PMC9136977
- DOI: 10.1021/acscentsci.1c01293
Synthetic Heparan Sulfate Mimetic Pixatimod (PG545) Potently Inhibits SARS-CoV-2 by Disrupting the Spike-ACE2 Interaction
Abstract
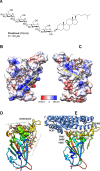
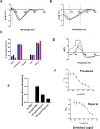
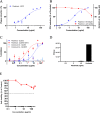
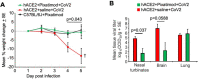
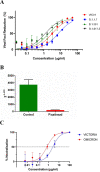
Heparan sulfate (HS) is a cell surface polysaccharide recently identified as a coreceptor with the ACE2 protein for the S1 spike protein on SARS-CoV-2 virus, providing a tractable new therapeutic target. Clinically used heparins demonstrate an inhibitory activity but have an anticoagulant activity and are supply-limited, necessitating alternative solutions. Here, we show that synthetic HS mimetic pixatimod (PG545), a cancer drug candidate, binds and destabilizes the SARS-CoV-2 spike protein receptor binding domain and directly inhibits its binding to ACE2, consistent with molecular modeling identification of multiple molecular contacts and overlapping pixatimod and ACE2 binding sites. Assays with multiple clinical isolates of SARS-CoV-2 virus show that pixatimod potently inhibits the infection of monkey Vero E6 cells and physiologically relevant human bronchial epithelial cells at safe therapeutic concentrations. Pixatimod also retained broad potency against variants of concern (VOC) including B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.2 (Delta), and B.1.1.529 (Omicron). Furthermore, in a K18-hACE2 mouse model, pixatimod significantly reduced SARS-CoV-2 viral titers in the upper respiratory tract and virus-induced weight loss. This demonstration of potent anti-SARS-CoV-2 activity tolerant to emerging mutations establishes proof-of-concept for targeting the HS-Spike protein-ACE2 axis with synthetic HS mimetics and provides a strong rationale for clinical investigation of pixatimod as a potential multimodal therapeutic for COVID-19.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): E. Hammond and K. Dredge are employees of Zucero Therapeutics. V. Ferro, E. Hammond, and K. Dredge are inventors on pixatimod patents.
Figures

References
-
- Cagno V.; Tseligka E. D.; Jones S. T.; Tapparel C. Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias?. Viruses 2019, 11, 596.10.3390/v11070596. - DOI
-
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

