BRAF-V600E utilizes posttranscriptional mechanisms to amplify LPS-induced TNFα production in dendritic cells in a mouse model of Langerhans cell histiocytosis
- PMID: 35648675
- PMCID: PMC9939017
- DOI: 10.1002/JLB.3A0122-075RR
BRAF-V600E utilizes posttranscriptional mechanisms to amplify LPS-induced TNFα production in dendritic cells in a mouse model of Langerhans cell histiocytosis
Abstract
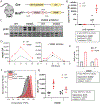
Langerhans cell histiocytosis (LCH) is an inflammatory disease characterized by abnormal dendritic cells (DCs) with hyperactive ERK signaling, called "LCH cells." Since DCs rely on ERK signaling to produce inflammatory molecules in response to pathogenic cues, we hypothesized that hyperactive ERK enhances DCs inflammatory responses. We specifically investigated TLR4-induced TNFα production in LCH cells by utilizing the BRAF-V600Efl/+ :CD11c-Cre mouse model of LCH, which hyperactivates ERK in DCs. We measured LPS-induced TNFα production both in vivo and in vitro using splenic CD11c+ cells and bone marrow-derived DCs with or without pharmacologic BRAFV600E inhibition. We observed a reversible increase in secreted TNFα and a partially reversible increase in TNFα protein per cell, despite a decrease in TLR4 signaling and Tnfa transcripts compared with controls. We examined ERK-driven, posttranscriptional mechanisms that contribute to TNFα production and secretion using biochemical and cellular assays. We identified a reversible increase in TACE activation, the enzyme required for TNFα secretion, and most strikingly, an increase in protein translation, including TNFα. Defining the translatome through polysome-bound RNA sequencing revealed up-regulated translation of the LPS-response program. These data suggest hyperactive ERK signaling utilizes multiple posttranscriptional mechanisms to amplify inflammatory responses in DCs, advancing our understanding of LCH and basic DC biology.
Keywords: cancer; dendritic cell; herbal medicines; immunosurveillance; innate immunity; signaling; translation.
©2022 Society for Leukocyte Biology.
Conflict of interest statement
DISCLOSURE
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Figures




References
-
- Favara BE, Feller AC, Pauli M, Jaffe ES, Weiss LM, Arico M, et al. Contemporary classification of histiocytic disorders. Med Pediatr Oncol. 1997;29:157–66. - PubMed
-
- van Furth R, Zanvil C. Origin and kinetics of mononuclear phagocytes. Ann N Y Acad Sci. 1976;278:161–75. - PubMed
-
- Morimoto A, Oh Y, Shioda Y, Kudo K, Imamura T. Recent advances in Langerhans cell histiocytosis. Pediatr Int. 2014;56:451–61. - PubMed
-
- Stålemark H, Laurencikas E, Karis J, Gavhed D, Fadeel B, Henter J-I. Incidence of langerhans cell histiocytosis in children: a population-based study helen. Pediatr Blood Cancer. 2008;50:1018–25. - PubMed
-
- Guyot-Goubin A, Donadieu J, Barkaoui M, Bellec S, Thomas C, Clavel J. Descriptive epidemiology of childhood langerhans cell histiocytosis in france, 2000–2004. Pediatr Blood Cancer. 2008;51:71–5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

